Viruses 2018 - Breakthroughs in Viral Replication
Part of the Viruses series
7–9 Feb 2018, Barcelona, Spain
- Go to the Sessions
- Event Details
-
- Welcome from the Chairs
- Sessions
- Instructions for Authors
- Event Gallery
- Sponsors and Partners
- Registration Fees
- Conference Venue
- Travel and Accommodation
- Scientific Committee
- Speakers
- Conference Awards
- Security Information
- Conference Secretariat
- Sponsoring Opportunities
- Conference Schedule
- Conference Dinner
- Events in this series
Welcome from the Chairs
Viruses 2018 - Breakthroughs in Viral Replication
![]() 7-9 February 2018
7-9 February 2018
![]() Faculty of Biology, University of Barcelona , Barcelona, Spain
Faculty of Biology, University of Barcelona , Barcelona, Spain
![]() Conference Awards 300€ & 500 €
Conference Awards 300€ & 500 €
Please feel free to download our Viruses 2018 poster and to help us to promote the conference in your Institute and University.
For sponsorship and exhibition opportunities please take a look at our brochure for Viruses 2018.
Welcome from the Conference Chair
Dear Colleagues,
It is with great pleasure that I announce the conference Viruses 2018 - Breakthroughs in Viral Replication to be held in Barcelona, Spain, 7-9 February 2018. The importance of viruses to human health has never been more apparent, and significant progress is being made in understanding virus replication, structure, transmission, pathogenesis, and antiviral immunity. This conference will bring together leading virologists from around the world to share their recent findings. Meeting participants will have the opportunity to present posters and short talks on their work and discuss their research in a relaxed, collegial environment.
The conference is sponsored by MDPI, the publisher of the open-access journal Viruses and follows the very successful meeting Viruses 2016 - At the Forefront of Virus-Host Interactions held in Jan 2016 in Basel, Switzerland.
I very much look forward to seeing you at this exciting meeting in Barcelona.
Best regards,
Eric O. Freed, Ph.D.
Editor-in-Chief, Viruses
Director, HIV Dynamics and Replication Program
Head, Virus-Cell Interaction Section
Center for Cancer Research
National Cancer Institute
Bg. 535/Rm. 110
Frederick, MD, USA
http://home.ncifcrf.gov/hivdrp/Freed.html
Conference Chair
 |
Dr. Eric O. FreedNational Cancer Institute, NIH - Frederick, MD, USA |
Conference co-Chair
 |
Dr. Albert BoschEnteric Virus Laboratory, School of Biology,
|
Confirmed Invited Speakers
|
|
|
|
|
Prof. Dr. Frank Kirchhoff |
Prof. Dr. Kay Grünewald |
Prof. Dr. Urs Greber |
|
|
|
|
|
Prof. Dr. Michael S. Diamond |
Dr. Alasdair Steven |
Dr. Alain Kohl |
|
|
|
|
|
Dr. Daniel Douek |
Dr. K. Andrew White |
Prof. Dr. Adam Zlotnick |
|
|
|
|
|
Dr. Sara Sawyer |
Prof. Dr. Clodagh O'Shea |
Dr. Elena I. Frolova |
|
|
|
|
|
Dr. Leo James |
Prof. Dr. Don C. Lamb |
Prof. Dr. Andrew B. Ward |
|
|
|
|
|
Dr. Amelia Nieto |
Prof. Dr. Ian Goodfellow |
Conference Secretariat
Sponsoring Opportunities
For information regarding sponsoring opportunities, please contact Antonio Peteira.
E-Mail: antonio.peteira@mdpi.com
Tel. +34 936397662
Instructions for Authors
The conference "Viruses 2018 - Breakthroughs in Viral Replication" will accept abstracts only.
- Create an account on Sciforum.net and follow the procedure to "Submit a New Abstract" in your User Home.
- Indicate which thematic area is best suited for your research.
- Submit an abstract in English - the word limits are minimum 150 words and maximum 300 words.
- The deadline to submit your abstract is 22 November 2017. You will be notified by 13 December 2017 about the acceptance for poster presentation.
- Upon submission, you can select if you wish to also be considered for oral presentation. Following assessment by the Chair, you will be notified by 13 December 2017 in a separate email whether your contribution has been accepted for oral presentation.
- Please note that, in order to finalize the scientific program in due time, at least one registration by anyone of the authors, denoted as Covering Author, is required to cover the presentation and publication of any accepted abstract. Covering Author registration deadline is 9 January 2018.
Maximum poster size limited to 90x140cm, vertical orientation preferred. Please print your poster prior to the conference.
A plan of the poster session will be circulated later on.
- All accepted abstracts will be available online in Open Access form on Sciforum.net.
- Participants of this conference are cordially invited to contribute with a full manuscript to our special issue "Breakthroughs in Viral Replication" in the journal Viruses.
- This themed collection is closely aligned with the scope of the event. The submission deadline for this special issue is
25 September 2018. The conference participants will be granted a 20% discount on the publishing fees. - Viruses is indexed by the Science Citation Index Expanded (Web of Science), MEDLINE (PubMed), and other databases, and has an Impact Factor of 3.465 (Journal Citations Reports, 2016).
Registration Fees
Registration closed: We have already reached the maximum number of participants. Thank you for your interest!
Registration Fees (in Euros)
| Early Registration: until 15 December 2017 | |
| Academic | 600 EUR |
| Section Editor and Guest Editor of Viruses* | 300 EUR |
| Scientific Board Members of Viruses | 450 EUR |
| MDPI Author** or Reviewer*** | 500 EUR |
| Student | 400 EUR |
| Non-academic | 1000 EUR |
| Late Registration: 16 December 2017 - 9 February 2018 - Closed |
|
| Academic | 700 EUR |
| Section Editor and Guest Editor of Viruses* | 300 EUR |
| Scientific Board Members of Viruses | 550 EUR |
| MDPI Author** or Reviewer*** | 600 EUR |
| Student | 400 EUR |
| Non-academic | 1100 EUR |
SEV Members (Sociedad Española de Virología) 350 EUR
The registration fee includes attendance of all conference sessions, morning/afternoon coffee breaks and lunches, participation in the conference dinner on the 7th of February, conference bag and program book. Other dinners are not included and are left at the convenience of attendees.
Please note that, in order to finalize the scientific program in due time, at least one registration by anyone of the authors, denoted as Covering Author, is required to cover the presentation and publication of any accepted abstract. Covering Author registration deadline is 9 January 2018.
* Guest Editors who served in 2015 or later will enjoy a discount.
** First and last authors of articles published in 2015 or later will enjoy a discount.
*** Reviewers who provided timely review reports in 2015 or later will enjoy a discount.
Cancellation Policy
Participation to the conference is considered final only once the registration fees have been paid. The number of participants is limited, once the number of paid registrations reaches the maximum number of participants, unpaid registrations will be cancelled.
| Cancellation of paid registration is possible under the terms listed below: | |
| > 2 months before the conference | Full refund but 100 EUR are retained for administration |
| > 1 month before the conference | Refund 50% of the applying fees |
| > 2 weeks before the conference | Refund 25% of the applying fees |
| < 2 weeks before the conference | No refund |
Conference Venue


Address: Avinguda Diagonal, 643, 08028 Barcelona, Spain
Closest Metro Station: Palau Reial (Line 3)
Barcelona
Barcelona is the capital and largest city of Catalonia and is Spain's second largest city, with a population of over one and half million people (over five million in the whole province).
This city, located on the northeastern Mediterranean coast of Spain, has a rich and diverse history, with its roots dating back to Roman times when it was a settlement called ‘Barcino’.
In 1992, Barcelona gained international recognition by hosting the Olympic games which brought about a massive upturn in its tourism industry.
This had the effect of changing the city in ways that are still felt today, with neighborhoods that were renovated (and in some cases, leveled), updates to the public transportation system, and an intense focus on modern design, that permeates all aspects of life in Barcelona, from public buildings to something as simple as a park bench or an event poster.
For visitors, this has translated into the very modern, yet incredibly old city you see now in the 21st century, where new elements work to both preserve and celebrate both the city’s heritage and origins.
Barcelona is similar to other large cosmopolitan European cities with plenty of outdoor markets, restaurants, shops, museums, and churches. The city is also very walkable, with an extensive and reliable Metro system for more far-flung destinations. The core center of town, focused around the Ciutat Vella ("Old City"), provides many opportunities for enjoyment for those looking to experience the life of Barcelona, while the beaches the city were built to provide locations for sun and relaxation during the long periods of agreeably warm weather.
For a complete overview, see wikitravel.org.



Travel and Accommodation
Barcelona, the capital of Catalonia, is one of the most popular tourist destinations in Europe. The city combines modern and historic architecture in a unique way. The city offers a wide range of accommodation so that everyone can find their perfect base camp by themselves.
| You can visit the official website of Barcelona Turisme for further information. |
Some reasons to choose Barcelona - More info
Accommodation
We want your stay in Barcelona during the conference to be as good as possible. With that in mind, we have reached an agreement with several hotels to make your accommodation choice a bit easier.
We are delighted to announce that the following hotel will offer a discounted price for all attendees to the Viruses 2018 conference:
- NH Collection Barcelona Constanza: Spacious, stylish rooms flooded with natural light, excellent connections with all Barcelona, and services designed for your comfort 10 minutes away from the conference venue by public transport.
Please use the following link to make your reservation: https://www.nh-hoteles.es/event/breakthroughs-in-viral-replication
Additionally, Atiram Hotels will offer 10% discount on room booking:
- Oriente Atiram and Meson Castilla Atiram: Located in Las Ramblas, have a fantastic central location, next to Plaza Catalunya and the famous Boqueria market, 25 minutes away (by metro) from the conference venue.
- Arenas Atiram: The Arenas Atiram Hotel offers a highly personalized service together with very friendly, elegant facilities, only a few meters away from the conference venue.
- Tres Torres Atiram: Located in the center of one of the best residential neighborhoods of Barcelona, next to the distinguished commercial and leisure districts Diagonal Avenue, 2 km away from the conference venue.
Please add the discount code ‘VIRUSES18’ while making your reservation.
Finally, the following hotels will also offer a 10% discount from 7 to 9 of Februray using the code ‘SERCOTELVIRUSES18’ :
- Hotel U232 offers great service and attention to detail and is located 20 minutes away by tram from the venue.
- B-Hotel is a modern hotel that offers comfort and design, located in Plaza España, 20 minutes away by metro from the conference venue.
- Hotel Barcelona Universal and Hotel Reding Croma are in an excellent location in the city centre 25 minutes away by metro from the venue.
To ensure availability, we encourage you to book your accommodation as soon as possible.
Getting to Barcelona
| By Plane |  |
By Train |  |
Once in Barcelona:
Public Transport (Bus & Metro): www.tmb.cat
Local Trains: www.fgc.cat
Some reasons to choose Barcelona - More info
Below are some references on trips and prices from Barcelona Airport to:
Faculty of Biology, University of Barcelona (Venue)
Working days 23 €. aprox.
Holidays, weekends and nights: 26 € aprox.
Plaza Catalunya (city center)
Working days: 26 € aprox.
Holidays, weekends and nights: 30 € aprox.
Public taxis can apply four types of fares in Barcelona. These fares must be visible inside the taxi, and are usually printed on a sticker over the window. Finally, the amount payable must be indicated on the taximeter.
Visa Requirements
Citizens of the EU and the EFTA
Citizens of EU and EFTA countries must present valid personal identification, an ID card or a passport. No visa is required.
Regardless of their citizenship, family members of EU and EFTA citizens must present a valid passport and they must have a specific residence permit from a Schengen country (no visa required). Otherwise a visa must be obtained.
For more information, see http://www.exteriores.gob.es/Portal/en/ServiciosAlCiudadano/InformacionParaExtranjeros/Paginas/Inicio.aspx.
EU Countries: Austria, Belgium, Bulgaria, Croatia, Cyprus, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Ireland, Italy, Latvia, Lithuania, Luxembourg, Malta, Netherlands, Poland, Portugal, Romania, Slovakia, Slovenia, Spain, Sweden, United Kingdom.
EFTA Countries: Iceland, Liechtenstein, Norway, Switzerland.
Schengen Countries: Austria, Belgium, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Italy, Latvia, Liechtenstein, Lithuania, Luxembourg, Malta, Netherlands, Norway, Poland, Portugal, Slovakia, Slovenia, Spain, Sweden, Switzerland.
Citizens of the USA and Canada
A valid passport is required. No visa is required for stays of up to 90 days.
Third-Country Nationals
A valid passport is required and in some cases a visa must be obtained prior to entering Spain. For more information, check http://www.exteriores.gob.es/Portal/en/ServiciosAlCiudadano/InformacionParaExtranjeros/Paginas/RequisitosDeEntrada.aspx.
Travel Insurance
Subscription to an insurance plan to protect you from the high costs of illness or accident during your stay in Spain is a prerequisite for obtaining your visa. You should obtain adequate travel, health and accident insurance before you depart from your country. The organizers are not responsible for personal injuries, or loss of, or damage to, private property belonging to the congress participants.
Letter of Invitation
A letter of invitation can be requested during the registration process using the "comments" field. The letter will be sent to you by email once the registration fees has been received. Letters of invitation do not include responsibility of, and, do not imply any financial support from the conference organizers.
Scientific Committee
Conference Chair
 |
Dr. Eric O. FreedNational Cancer Institute, NIH - Frederick, MD, USA |
Conference co-Chair
 |
Dr. Albert BoschEnteric Virus Laboratory, School of Biology,
|
Scientific Advisory Committee Members
 |
 |
 |
| Dr. Sandra K. Weller University of Connecticut, USA |
Dr. Carolyn Coyne University of Pittsburgh, USA |
Dr. Sara Cherry |
 |
 |
| Dr. Ralf Bartenschlager University of Heidelberg, Germany |
Dr. Felix A. Rey Pasteur Institute, France |
Speakers
Confirmed Invited Speakers
|
|
|
|
|
Prof. Dr. Frank Kirchhoff |
Prof. Dr. Ian Goodfellow |
Prof. Dr. Urs Greber |
|
|
|
|
|
Prof. Dr. Michael S. Diamond |
Dr. Alasdair Steven |
Dr. Alain Kohl |
|
|
|
|
|
Dr. Daniel Douek |
Dr. K. Andrew White |
Prof. Dr. Adam Zlotnick |
|
|
|
|
|
Dr. Sara Sawyer |
Prof. Dr. Clodagh O'Shea |
Dr. Elena I. Frolova |
|
|
|
|
|
Dr. Leo James |
Prof. Dr. Don C. Lamb |
Prof. Dr. Andrew B. Ward |
|
|
|
|
|
Dr. Amelia Nieto |
Conference Awards
The Sponsor, Viruses, offers two awards to our participants at the conference:
1. Best Poster Award (1): 300€.
2. Best Oral Presentation Award (1): 500€
The nominations will be assessed by the Organizing Committee.
Security Information
As you may know, as a response to the current political situation in Spain and Catalonia, there have been peaceful protest movements occurring in Barcelona. We do not expect any related disruption at the conference site and there should be no impact on the meeting. Moreover, the demonstration up till now have been completely peaceful and have not caused us to be concerned regarding our safety/security. Finally, during the last months we have successfully held three other conferences in Barcelona (two in September and one in October) and we are working to make Viruses 2018 a great meeting for the Virology community.
The Viruses 2018 Organization Team
Conference Secretariat
Sponsoring Opportunities
E-Mail: antonio.peteira@mdpi.com
Tel. +34 936397662
Conference Schedule
The detailed conference program of Viruses 2018 - Breakthroughs in Viral Replication is now available, please see below.
Also, the book of abstracts of the conference can be downloaded here (last update 30th May 2019).
Program Structure - Viruses 2018: Breakthroughs in Viral Replication, 7-9 February 2018, Barcelona (Spain) |
|||
|
Wednesday 7 February 2018 |
Thursday 8 February 2017 |
Friday 9 February 2018 |
|
|
Morning |
Check-in Program Overview and Introduction Chairs: Eric O. Freed and Albert Bosch S1. General Topics in Virology |
S3. Virus Replication |
S5. Genome Packaging and Replication/Assembly |
|
Coffee Break |
|||
|
S1. General Topics in virology |
S3. Virus Replication |
S5. Genome Packaging and Replication/Assembly |
|
|
Lunch |
|||
|
|
S2. Structural Virology |
S4. Replication and Pathogenesis of RNA Viruses |
S6. Antiviral Innate Immunity and Viral Pathogenesis |
|
Coffee Break |
Apéro and Poster Session Conference Group Photograph |
Coffee Break |
|
|
S2. Structural Virology |
S6. Antiviral Innate Immunity and Viral Pathogenesis Closing Remarks |
||
| Conference Dinner | |||
Session Start
Wednesday 7 February 2018: 08:00 - 12:30 / 14:00 - 18:00 / Conference Dinner: 20:30
Thursday 8 February 2018: 08:30 - 12:30 / 14:00 - 18:30
Friday 9 February 2018: 08:30 - 12:30 / 14:00 - 18:15
Detailed Program
Day 1: Wednesday 7 February 2018
08:00 – 08:45 Check-in
08:45 – 09:00 Program Overview and Introduction - Eric O. Freed and Albert Bosch
Session 1: General Topics in Virology - Session Chairs: Thomas Klimkait and Joanna Parish
09:00 – 09:30 Ralf Bartenschlager: "New insights into Flaviviridae – host cell interactions"
09:30 – 09:45 Rachel Whitaker: "CRISPR-Cas co-evolutionary dynamics in natural host-virus populations"
09:45 – 10:00 Michael Bauer: "Ubiquitination-dependent Adenovirus Capsid Disassembly at the Nuclear Pore Complex"
10:00 – 10:15 Moriah Szpara: "Genomic and phenotypic diversity associated with neonatal HSV-2 disease"
10:15 – 10:45 Coffee Break
10:45 – 11:15 Rosa Pintó: "Hepatitis A virus codon usage: implications for virus biology and phenotype"
11:15 – 11:45 Michael S. Diamond: "CRISPR-Cas9 Screens Define Novel Host Factors Required for Infection by Arthritogenic Alphaviruses"
11:45 – 12:00 Itay Rousso: "Reverse transcription mechanically induces HIV-1 capsid disassembly"
12:00 – 12:15 Heinrich Gottlinger: "SERINCs and HIV Infectivity"
12:15 – 12:30 Inés Romero Brey: "Visualizing the 3D architecture of the Hepatitis A Virus replication organelles"
12:30–14:00 Lunch
Session 2: Structural Virology - Session Chairs: Delphine Muriaux and Kay Choi
14:00 – 14:15 Yohei Yamauchi: "An Influenza Uncoating Epitope Confers Capsid Stability"
14:15 – 14:45 Felix A. Rey: "Relations between Cell–Cell Fusion, Eukaryotic Evolution and Class II Enveloped Viruses Identified by Structural Studies"
14:45 – 15:00 Kelly Lee: "Dynamics bridge structure, function, and phenotype in viral entry machines"
15:00 – 15:15 Hadas Cohen Dvashi: "Structure-based rational design of broad range immunotherapy targeting TfR1-tropic arenaviruses"
15:15 – 15:30 Matteo Castelli: "Unraveling the structure-function relationship of hepatitis C virus fusion machinery by in silico structural modeling"
15:30 – 16:00 Coffee Break
16:00 – 16:30 Eric O. Freed: "HIV-1 Assembly, Release, and Maturation"
16:30 – 17:00 Leo James: "Viral genome hide and seek; encapsidation versus host immunity"
17:00 – 17:15 Ana Joaquina Perez-Berná: "Correlative study of cellular modification induced by Hepatitis C infection by cryo soft X-ray tomography and Infrared microscopy"
17:15 – 17:30 Diego Ferrero: "Supramolecular arrangement of the Zika virus NS5 protein"
17:30 – 17:45 Jamil Saad: "Towards elucidating the structural basis for envelope incorporation into HIV-1 particles"
20:30 Conference Dinner
Day 2: Thursday 8 February 2018
Session 3: Virus Replication Compartments - Session Chairs: Stefano Aquaro and Nathalie Arhel
08:30 – 09:00 Sandra Weller: "Roles of UL12 and ICP8 in the production of Herpes Simplex Virus DNA that can be packaged into infectious virus"
09:00 – 09:30 Sara Sawyer: "Spillover: How viruses adapt at the animal - human interface"
09:30 – 09:45 Thejaswi Nagaraju: "Mapping Epstein-Barr viral genome replication spatially and temporally during the viral lytic cycle"
09:45 – 10:00 Jean-Francois Eleouet: "Respiratory syncytial virus RNA synthesis in cytoplasmic inclusion bodies: organization and functioning"
10:00 – 10:15 Griffith Parks: "Cytoplasmic-Replicating Parainfluenza Virus Sensitizes the Nuclear Compartment to DNA-Damaging Agents; Implications for Oncolytic Virus Therapies"
10:15 – 10:45 Coffee Break
10:45 – 11:15 Clodagh O’shea: "Cracking Nuclear Codes: The 5D viral-cellular genome structures and interactions that drive pathological proliferation"
11:15 – 11:45 Elena I. Frolova: "Assembly of viral replication complexes: Roles for viral and host proteins"
11:45 – 12:00 Luis DaSilva: "ESCRT machinery activity is required for oropouche virus assembly at the Golgi complex"
12:00 – 12:15 Montserrat Bárcena: "Common themes in the ultrastructure and function of the coronavirus replication organelle"
12:15 – 12:30 Catherine Eichwald: "Core or Viroplasm? Dissecting rotavirus VP2 and NSP5 interaction"
12:30–14:00 Lunch
Session 4: Replication and Pathogenesis of RNA viruses - Session Chairs: Josep Quer and Richard Sutton
14:00 – 14:30 Amelia Nieto: "Epigenetic control of influenza virus: role of H3K79 methylation in interferon-induced antiviral response"
14:30 – 15:00 Ian Goodfellow: "Identification of trans acting factors involved in the norovirus life cycle."
15:00 – 15:15 Rachel van Duyne: "HIV-1 envelope glycoproteins confer broad resistance to antiretrovirals in vitro"
15:15 – 15:30 Natalya Teterina: "MicroRNA-based suppression of neurotropic flavivirus replication: application for the development of live attenuated vaccine"
15:30 – 15:45 Mariana Batista: "Adaptation of NrHV to the mouse host using selective innate and adaptive KO strains"
15:45 – 16:00 Fernando Ponz: "Differential developmental symptoms in Turnip mosaic virus-infected hosts is associated with the dynamic association of the viral P3 protein to the endoplasmic reticulum"
16:00 – 16:15 Pietro Scaturro: "An Orthogonal Proteomic Screen of Zika virus Uncovers Novel Host-dependency Factors"
16:15 – 16:30 Conference Group Photograph
16:30 – 18:30 Apéro and Poster Session
Day 3: Friday 9 February 2018
Session 5: Genome Packaging and Replication/Assembly - Session Chairs: Juan José López-Moya and François-Loïc Cosset
08:30 – 09:00 K. Andrew White: "Intra-genomic RNA-based Regulation in a Plus-strand RNA Virus"
09:00 – 09:30 Adam Zlotnick: "Assembly and disassembly of HBV: biology recapitulates physics"
09:30 – 09:45 Kay Choi: "Crystal structure of the dengue virus promoter RNA"
09:45 – 10:00 Vibhu Prasad: "Adenovirus activation of the unfolded protein response sensor Ire1 enhances immediate early viral transcription to promote viral persistence and lytic egress"
10:00 – 10:15 Myra Hosmillo: "Identification of cis-acting RNA sequences involved in norovirus VPg-dependent RNA synthesis"
10:15 – 10:45 Coffee Break
10:45 – 11:15 Andrew Ward: "Structure, function, and immune recognition of the enigmatic HIV envelope glycoprotein" - Young Investigator Award Winner 2017
11:15 – 11:30 Alexander Borodavka "Protein-Assisted RNA Folding Mediates Specific RNA–RNA Genome Segment Interactions in Segmented RNA Viruses"
11:30 – 11:45 Brett Lindenbach: "The hepacivirus and pestivirus NS3 helicases act as motor proteins to power RNA encapsidation during virus particle assembly"
11:45 – 12:00 Solène Denolly: "The Amino-Terminus of the Hepatitis C Virus (HCV) p7 Viroporin and Its Cleavage from Glycoprotein E2-p7 Precursor Determine Specific Infectivity and Secretion Levels of HCV Particle Type"
12:00 – 12:15 José Almendral: "Differential Phosphorylation and N-terminal Configuration of Capsid Subunits in Parvovirus Assembly and Viral Trafficking"
12:15–14:00 Lunch
Session 6: Antiviral Innate Immunity and Viral Pathogenesis - Session Chairs: Shan-Lu Liu and Eric Poeschla
14:00 – 14:30 Carolyn Coyne: "Stem Cell-derived Enteroids to Model Enterovirus-GI Interactions"
14:30 – 15:00 Sara Cherry: "Harnessing genomic approaches to explore the interface between viruses and hosts"
15:00 – 15:30 Urs Greber "Towards Understanding ‘Cell-to-Cell’ Variability of Viral Gene Expression"
15:30 – 15:45 Eric Snijder: "Middle East respiratory syndrome-coronavirus employs the deubiquitinating activity of its nsp3 papain-like protease to suppress the innate immune response"
15:45 – 16:15 Coffee Break
16:15 – 16:45 Frank Kirchhoff "HIV Accessory Factors: More Than One Way to Skin a Cat"
16:45 – 17:15 Alain Kohl "Arboviruses and antiviral RNAi pathways in mosquito cells: progress in understanding regulation and effectors"
17:15 – 17:30 Susana Guerra: "ISG15 governs mitochondrial function in macrophages following vaccinia virus infection"
17:30 – 17:45 Pilar Domingo-Calap: "Virus-virus interactions driven by innate immune responses"
17:45 – 18:00 John Schoggins: "Interferon inhibits flavivirus replication via IFI6, a chaperone-dependent ER membrane effector"
18:00 – 18:15 Closing Remarks
Conference Dinner
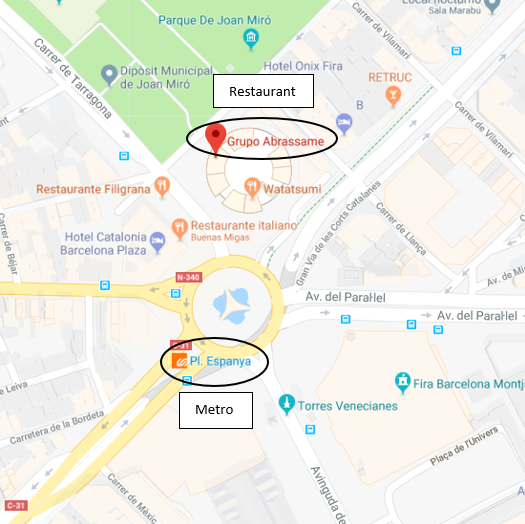
We are glad to inform that the conference dinner will be held at Abrassame, a cutting-edge restaurant specialised in Mediterranean cuisine which, in addition to its location at the pictouresque terrace of Arenas de Barcelona, will make of your evening at the restaurant an experience to remember.

Abrassame is located at Gran Via de les Corts Catalanes 373 – 385, only a few minutes away from one of one of Barcelona’s most emblematic must-sees, Montjüic. You can easily reach the restaurant from the conference venue either by taxi or by Metro. If you were to choose the second option, the easiest way to get there is by taking the Metro Line L3 at Palau Reial Station and drop off at Espanya Metro Station after 6 stops.
S1. General Topics in Virology
Confirmed Speakers
University of Heidelberg, Germany
Dr. Rosa Pintó
University of Barcelona, Spain
Prof. Dr. Michael S. Diamond
Washington University in St. Louis, School of Medicine, USA
Show all accepted abstracts (32) Hide accepted abstracts (32)
List of Accepted Abstracts (32) Toggle list
S2. Structural Virology
Confirmed Speakers
Dr. Alasdair Steven
National Institutes of Health (NIH), USA
Dr. Félix A. Rey
Institut Pasteur, France
Dr. Leo James
Medical Research Council (MRC), UK
Prof. Dr. Andrew B. Ward - Young Investigator Prize recipient
The Scripps Research Institute, USA
Session Chair
Professor Felix Rey
Show all accepted abstracts (19) Hide accepted abstracts (19)
List of Accepted Abstracts (19) Toggle list
S3. Virus Replication Compartments
Confirmed Speakers
University of Connecticut, USA
Dr. Sara Sawyer
University of Colorado Boulder, USA
Salk Institute for Biological Studies, USA
Dr. Elena I. Frolova
UAB School of Medicine, USA
Session Chair
Professor Ralf Bartenschlager
Show all accepted abstracts (21) Hide accepted abstracts (21)
List of Accepted Abstracts (21) Toggle list
S4. Replication and Pathogenesis of RNA viruses
Confirmed Speakers
Spain
Dr. Ian Goodfellow
University of Cambridge, UK
Session Chair
Dr. Albert Navarro
Show all accepted abstracts (45) Hide accepted abstracts (45)
List of Accepted Abstracts (45) Toggle list
S5. Genome Packaging and Replication/Assembly
Confirmed Speakers
York University, Canada
Prof. Dr. Adam Zlotnick
Indiana University, USA
Dr. Eric O. Freed
National Cancer Institute, NIH, USA
Ludwig-Maximilians-Universität München, Germany
Session Chairs
Dr. Carolyn Coyne, University of Pittsburgh
Dr. Sandra Weller
Show all accepted abstracts (31) Hide accepted abstracts (31)
List of Accepted Abstracts (31) Toggle list
S6. Antiviral Innate Immunity and Viral Pathogenesis
Confirmed Speakers
University of Pittsburgh, USA
Dr. Sara Cherry
University of Pennsylvania, USA
Prof. Dr. Urs Greber
Institute of Molecular Life Sciences, University of Zurich, Switzerland
Ulm University Hospital, Germany
Dr. Alain Kohl
University of Glasgow, UK
Session Chair
Professor Sara Cherry
Show all accepted abstracts (44) Hide accepted abstracts (44)
List of Accepted Abstracts (44) Toggle list