
The 3rd International Electronic Conference on Cancers: New Targets for Cancer Therapies (IECC 2023)
Part of the International Electronic Conference on Cancers (IECC) series
16–30 March 2023
Cancer, Cancer Genomics, Epigenomics, Cancer Epidemiology, Cancer Research
- Go to the Sessions
- Event Details
The recordings of IECC 2023 live sessions are available at: https://iecc2023.sciforum.net/#recordings
The IECC 2023 award winners have been announced at https://iecc2023.sciforum.net/#awards
Registration for IECC 2023 live sessions is open now
There will be two live sessions, all of which are completely free to attend. There is a different registration link for each live session. You only need to register for the ones that you wish to join. If you would like to attend more than one live session, please register for each session separately.
Registrations with academic institutional email addresses will be prioritized.
- 20 March 2023 at 3:00pm CET (BASEL TIME) --> Registration Link: https://us06web.zoom.us/webinar/register/1416752495484/WN_NulyJDYsQyatCWUoUPkutg
- 28 March 2023 at 3:00pm CEST (BASEL TIME) --> Registration Link: https://us02web.zoom.us/webinar/register/6116752497021/WN_oHhZXvImQKCXfCrMvPCqpQ
Welcome from the Chair
Dear colleagues,
We are pleased to announce the 3rd International Electronic Conference on Cancers: New Targets for Cancer Therapies (IECC 2023), which will be held from 16–30 March 2023.
The opportunities for new therapeutic targets have expanded tremendously. In recent years, cancer genomics has expanded our view of cancer vulnerabilities, new technologies have changed the paradigm of what can be successfully targeted, and new immunotherapy targets are emerging rapidly. This conference aims to promote and advance the coordinated progress in oncology research to foster the development of innovative therapeutic strategies. It will bring together experts in basic, translational, and clinical research to discuss the current research and the opportunities as well as obstacles that lie ahead in the field.
The focus of this conference is the translation of recent discoveries into new therapeutic targets for cancer therapies. Topics of interest include, but are not limited to, the following:
- Translational cancer genomics and epigenomics.
- Emerging immunotherapy targets.
- Overcoming therapeutic hurdles in resistance and undruggable proteins.
There will be five specific sessions:
Session 1: Genome-wide Approaches for Target Identification.
Session 2: Targeting Metastasis.
Session 3: Novel Immunotherapy Targets.
Session 4: Novel Approaches for ‘Undruggable’ Targets.
Session 5: Overcoming Therapeutic Resistance.
Participants will have the opportunity to examine, explore, and critically engage with issues and advances in these areas. We hope to facilitate discussions and exchange within the community.
This event will solely be an online proceeding, which allows for participation from all over the world with no concerns of travel and related expenditures. This type of conference is particularly appropriate and useful because research concerned with cancers is progressing rapidly. An electronic conference provides a platform for rapid and direct exchanges about the latest research findings and novel ideas. Participation in, as well as the “attendance” of, this online conference is free of charge.
This electronic conference is sponsored by MDPI and the scientific journal Cancers. The accepted conference proceedings papers and presentations will be available online for discussion during 16–30 March 2023 and will be published in the journal Medical Sciences Forum. All participants are cordially encouraged to submit an extended full manuscript to the Cancers conference Special Issue "IECC2023: New Targets for Cancer Therapies" with a 20% discount on the APC. Cancers (ISSN 2072-6694; IF 6.575) is a peer-reviewed and open access journal of oncology that is published semimonthly online by MDPI.
The best paper will receive an award of CHF 500, and the best presentation (poster, slides, or video) will receive an award of CHF 300. Both will also receive an offer to publish an extended paper, with a 20% discount, in a Special Issue "IECC2023: New Targets for Cancer Therapies" of the journal Cancers.
We hope that the community will share this enthusiasm and help make this conference a success.
Kind regards,
Dr. Carlos S. Moreno
The Chair of the 3rd International Electronic Conference on Cancers: New Targets for Cancer Therapies
Follow the conference organizer on Social Media
Live Session Program
Live Session 1
20 March 2023
3:00 pm CET | 10:00 am EDT | 10:00 pm CST Asia
Session Chair: Prof. Dr. Nicola Amodio
|
Speaker |
Presentation Topic |
Time (CET) |
|
Prof. Dr. Nicola Amodio |
Introduction |
3:00pm-3:10pm |
|
Dr. Andrea Morandi |
Lipid metabolism is a vulnerability of endocrine-resistant breast cancers |
3:10pm–3:40pm |
|
Dr. Mariateresa Fulciniti |
Targeting myeloma at the intersection between cell cycle, metabolism and transcriptional regulation to repress growth and overcome resistance |
3:40pm-4:10pm |
|
Prof. Dr. Nicola Amodio |
Conclusion |
4:10pm-4:20pm |
Live Session 2
28 March 2023
3:00 pm CEST | 9:00 am EDT | 9:00 pm CST Asia
Session Chair: Dr. Francisca Vazquez
|
Speaker |
Presentation Topic |
Time (CET) |
|
Dr. Francisca Vazquez |
Introduction |
3:00pm-3:10pm |
|
Dr. Francesco Iorio |
Optimisation and drug-discovery oriented analyses of CRISPR-Cas9 screens |
3:10pm-3:40pm |
|
Dr. Travert Hart |
Multiplex perturbation strategies for genetic modeling of drug response |
3:40pm-4:10pm |
|
Dr. Davide Barbagallo |
The complex crosstalk among non-coding RNAs: the case of circSMARCA5 in glioblastoma multiforme |
4:10pm-4:40pm |
|
Prof. Dr. Vasso Apostolopoulos |
Identification of new cancer markers: a way forward |
4:40pm-5:10pm |
|
Dr. Francisca Vazquez |
Conclusion |
5:10pm-5:20pm |
Live Session Recordings

Event Chair

Department of Pathology and Laboratory Medicine, Emory University School of Medicine, USA
Carlos S. Moreno, PhD, is an Associate Professor in the Departments of Pathology and Laboratory Medicine, and Biomedical Informatics at the Emory University School of Medicine. He is a member of the Cell and Molecular Biology Research Program at Winship Cancer Institute. Dr. Moreno specializes in cancer bioinformatics and systems biology, analysis of genome-wide expression profiles, transcriptional networks, ChIP-chip studies, biological pathway analysis, and computational analysis of transcription factor binding sites. Dr. Moreno is project leader for informatics for the Emory Molecular Interaction Center for Functional genomics (MicFG) as part of the Cancer Target Discovery and Development (CTD²) Network to analyze TCGA data for protein-protein interaction networks.
cmoreno@emory.edu
Session Chairs

Prof. Dr. Frank McCormick
UCSF Helen Diller Family Comprehensive Cancer Center, School of Medicine, University of California, USA; NCI RAS Initiative, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, USA
Frank McCormick, PhD, FRS, is a Professor in the UCSF Helen Diller Family Comprehensive Cancer Center. Prior to joining the UCSF faculty, Dr. McCormick pursued cancer-related work with several Bay Area biotechnology firms and held positions with Cetus Corporation and Chiron Corporation, where he was Vice President of Research from 1991 to 1992. Dr. McCormick holds the David A. Wood Chair of Tumor Biology and Cancer Research at UCSF. Dr. McCormick is the author of over 285 scientific publications and holds 20 issued patents. He also served as President, 2012-2013 for the American Association for Cancer Research (AACR). More recently, he has taken a leadership role at the Frederick National Lab for Cancer Research, overseeing an NCI supported national effort to develop therapies against Ras-driven cancers. These cancers include most pancreatic cancers, and many colorectal and lung cancers, and are amongst the most difficult cancers to treat.

Dr. Francisca Vazquez
Broad Institute of MIT and Harvard, USA
Francisca Vazquez is the director of the Cancer Dependency Map Project (DepMap) at the Broad Institute, a programmatic effort that she co-leads to systematically identify all cancer dependencies in human cancer. The DepMap project is an effort to comprehensively identify genetic dependencies and small molecules sensitivities and the molecular markers that predict their response across thousands of cancer cell line models to accelerate precision cancer medicine. To accomplish this goal, Vazquez works with a large interdisciplinary team of project managers, basic scientists, and computational biologists that uses state-of-the-art genome-scale functional genomics, including CRISPR and high-throughput small molecule screening technologies, to systematically profile hundreds of cellular models of cancers. Vazquez also leads DepMap’s target discovery and advancement team, which focuses on developing methods to systematically identify the most promising therapeutic targets and translate the findings to therapeutics. Vazquez launched the Pediatric and Brain Tumor dependency maps, which she co-leads to more rapidly advance therapeutic discoveries in those tumor types. Her main goal is to help accelerate precision cancer medicine by both creating resources for the scientific community, such as DepMap, and identifying and validating the most promising targets and biomarkers. Prior to joining the Broad Institute in 2011, Vazquez worked as a research scientist at Dana-Farber Cancer Institute in the center for chronic disease, studying cancer metabolism; her postdoctoral work, which focused on tumor suppressor genes, was also undertaken at DFCI. She earned her Ph.D. from the University of Illes Balears in Spain, while performing most of her work as a visiting student at Beth-Israel Deaconess Medical Center.

Prof. Dr. Nicola Amodio
Department of Experimental and Clinical Medicine, Magna Graecia University of Catanzaro, Italy
Nicola Amodio, Ph.D., joined the University Magna Graecia of Catanzaro (I) in 2002 as a PhD fellow in “Molecular Oncology”, focusing on transcription factors dysregulation in normal and malignant hematopoiesis. After a Post-Doctoral fellowship at MSKCC (NY, USA) in 2006, he returned at University of Catanzaro as a Senior Scientist, focusing on the discovery of genetic and epigenetic alterations driving the development of solid and hematologic malignancies, and on the design of innovative anti-cancer therapies. Since 2019, Dr. Amodio works as Assistant Professor, and since 2022 he has been appointed Associate Professor of “General Pathology” at the Department of Experimental and Clinical Medicine, University Magna Graecia of Catanzaro (I). He serves as editor in chief for the journal “Cancers”, section “cancer pathophysiology”. His current research, supported by the Italian Association for Cancer Research (AIRC), is aimed at the identification and characterization of novel druggable vulnerabilities underlying the pathogenesis and progression of plasma cell dyscrasias by using integrated functional and (epi)genomics analyses.
Event Committee

Program of Immunology and Immunotherapy. Cima Universidad de Navarra, Spain
Pedro Berraondo, Ph.D., is a staff scientist at the Cima Universidad de Navarra. He is a co-inventor of eight patents, and co-author of more than 140 publications including publications in Nature, Nature Medicine, Nature Reviews Drug Discovery, Immunity, Cancer Discovery, and Cancer Cell. He is co-director of the degree of an expert in immuno-oncology at the Faculty of Medicine of the University of Navarra, and the coordinator of the immuno-oncology work module at CIBERONC.
Cancer immunotherapy; fusion proteins; gene therapy

Dr. Avila is Professor of Biochemistry and Molecular Biology and director of the Hepatology Program of the Center for Applied Medical Research at the University of Navarra, Pamplona, Spain. His research interests include the cellular and molecular mechanisms of the hepatic response to acute injury and regeneration, as well as the genetic mechanisms of chronic liver disease progression and carcinogenesis, with a basic and translational perspective. Research for the identification of novel biomarkers for the early diagnosis of hepatobiliary malignancies in the context of liquid biopsy is also pursued in his laboratory.
hepatocarcinogenesis; cell signaling; differentiation; epigenetics

Department of Agricultural Sciences University of Naples Federico II, Italy
Degree in Parmacy (1996); Master in Medicinal Plants (2000); Master in Pharmacology (2002); PhD in Drug Science (2005). TEACHING EXPERIENCE: Professor of Pharmacognosy at the University of Naples Federico II. MAIN RESERCH INTERESTS: Role of cannabinoid, vanilloid and kappa-opioid receptors in the gastrointestinal tract, both in physiological (e.g. intestinal motility) and pathophysiological states (e.g. inflammation and cancer); Ethnopharmacological studies on medicinal plants and their active ingredients used in traditional medicine; Clinical pharmacology of herbal products; Nutritional pharmacology. PUBLICATIONS: 190 articles in peer-reviewed international Journals (cited in JCI). AWARDS: Farmindustria Prize (year 2006), from the Italian Society of Pharmacology, to the best article published by researcher under 35 years as a first author.
pharmacology; natural products; neurotransmission; behavioral pharmacology; experimental pharmacology; preclinical pharmacology; CB1 receptor; PPARs; cannabinoids; endocannabinoids; CB2 receptor

Department of Experimental and Clinical Biomedical Sciences, Experimental Pathology and Oncology, University of Florence, Italy
Francesca Bianchini is a tenured associate professor in General Pathology at the University of Florence involved in teaching to Medicine and Surgery, and Nursing students. After graduating in Biological Sciences, she received her training on preclinical oncology, under the supervision of Prof. Calorini, at the University of Florence. She obtained her PhD in Experimental Pathology from the University of Florence in 2002. During her PhD, she attended the laboratory of Prof. Charles N. Serhan at the B&WH Harvard Medical School in Boston (MA). Her post-doctoral research has been extensively focused on preclinical oncology and translational medicine.
tumor microenvironment (TME); cancer cell biology; extracellular acidity; nanomedicine; targeted drug delivery and personalized anticancer therapy

Cancer Biomarkers Unit, Fondazione IRCCS Casa Sollievo della Sofferenza, Italy
He worked as postdoctoral fellow at the FIRC Institute for Molecular Oncology (IFOM), Milan (Italy), and then at the Program of Molecular Medicine of the European Institute of Oncology (Milan). He was a visiting scientist at the Nuffield Department of Clinical and Laboratory Sciences at University of Oxford. Since 2016 he has been the Head of the Cancer Biomarkers Unit at the Research Hospital (IRCCS) Casa Sollievo della Sofferenza. He has several years of research experience in the fields of cancer genomics and biomarkers development, particularly in the field of circulating miRNAs, cancer gene expression profile and computational biology. He has pioneered the identification of serum circulating miRNA-signature for lung cancer early detection. He is inventor of international patents regarding diagnostic and prognostic tools for lung cancer screening. He is an active member of the American Association for Cancer Research (AACR), the European Association for Cancer Research (EACR), the International Association for the Study of Lung Cancer (IASLC), the Italian Society for Cancer Research (AIRC), and the Italian Society of Cancerology (SIC), where he holds the positions of secretary and member of the steering committee.
lung cancer; molecular oncology; gene expression; cancer biomarkers; microRNA; exosomes

Department of Internal Medicine-DIMED, Internal Medicine and Hepatology Unit, University-Hospital AOUP, Italy;
Responsible of the Out-patient Unit of the Veneto Specialized Center on Liver Diseases, AOUP, Italy
- Degree in Medicine and Surgery at the University of Padua (UNIPD), Italy on 03/28/83 - Qualification as a Surgeon in the 1st 1983 session with registration N°5037 on 05/07/1983 to the Order of Physicians and Surgeons of Padua, Italy. - Specialization in Internal Medicine at UNIPD on 07/15/88. - Graduate Technical Assistant at the UNIPD-Institute of Clinical Medicine from 1991 to 1997. - Level I Medical Director in Internal Medicine-Hospital of Padua since 06/1991. - PhD in Clinical Pharmacology and Medical Therapy at UNIPD in 1993. - Confirmed Researcher and Adjunct Professor from 2006 to 2015 and - Confirmed Associate Professor of Internal Medicine from 10/01/2015 of the Department of Medicine-DIMED at UNIPD.
clinical and epidemiological features of chronic hepatitis C and B and its antiviral treatments; cirrhosis and complication of evolutive liver disease; natural history and treatment of hepatocellular carcinoma; transient elastography for staging of chroni

Dr. Aamir Ahmad is a Senior Research Scientist and Group Leader at Academic Health System and Dermatology Institute, Translational Research Institute, Hamad Medical Corporation (HMC). Before joining HMC, he was a Senior Research Scientist at University of Alabama at Birmingham, Birmingham, AL, USA. Dr. Ahmad holds a PhD degree from AMU, Aligarh, India and completed his post-doctoral training at National Cancer Institute, National Institutes of Health, Bethesda, MD, USA. He has authored more than 200 peer-reviewed publications (h-index: 68) and 30 book chapters, in addition to editing books for leading publishing houses and serving on the editorial board of several International Journals of repute. Dr. Ahmad’s research focuses on epigenetic regulation of disease progression, particularly cancer and skin diseases.
cancer epigenetics; metastasis; cancer drug resistance; tumor microenvironment; miRNA; non-coding RNAs; cancer stem cells

Department of Pathology, Department of Anatomy & Cell Biology, Rush University Medical Center, USA
Jeffrey A. Borgia, PhD. is a graduate of the Biochemistry Graduate program at the University of Minnesota where he performed research in the field of proteoglycan biosynthesis in the laboratory of Dr. Theodore R. Oegema, Jr. During his post-doctoral studies, he collaborated with Dr. Gregg B. Fields where he explored the production of matrix metalloproteinases by melanoma cells when adhered to synthetic ‘mini-collagens’ as well as tumor cell aggressiveness. In 2002, Dr. Borgia joined the faculty at Rush in the Department of Biochemistry and launched his program focused on cancer biology and the development of novel diagnostic tests with Dr. John S. Coon, IV in the Department of Pathology. He is currently continuing these studies with appointments in both the Department of Cell & Molecular Medicine and Department of Pathology.
lung cancer; biomarkers; cancer cachexia; immunotherapy; recurrence; lung cancer screening; early detection; EMT; proteomics; biobanking

His team is working on RNA metabolism, including alternative splicing, and on the role of RNA metabolism in the phenotypic plasticity of cancer cells, including in response to anti-cancer therapies. His expertise is notably based on the use of original bioinformatics analysis approaches.
Splicing; Splicing Factors; RNA Binding Proteins; RNA Processing.

Instituto de Investigaciones Biomédicas Alberto Sols (CSIC-UAM), Arturo Duperier
Lisardo Boscá graduated with a Ph.D. in sciences from the University of Autónoma de Madrid. In the last decade, Dr. Lisardo Boscá has been working in the regulation of inflammation as a common pathophysiological process, from inception to resolution, and applied this knowledge to understand the pathogenesis of processes that are relevant in cardiovascular diseases. In 2010, in a paper published in J. Immunol., with more than 600 citations, Dr. Lisardo Boscá described the metabolic features associated with the phenotypic differentiation of macrophages, from a pro-inflammatory to an anti-inflammatory and pro-resolution profile. In addition to this, Dr. Lisardo Boscá has shown for the first time how bioactive lipids derived from cyclooxygenase and/or lipooxygenases are involved in the resolution of several cardiovascular pathologies, from atherothrombosis to myocardial infarction to autoimmune myocarditis.
cancer immunometabolism; cancer therapeutics; apoptosis; immunotherapy; cancer metabolism; colorectal cancer

INSERM UMR 1186, Integrative Tumor Immunology and Genetic
Oncology, Gustave Roussy, EPHE, Fac. de médecine-Univ. Paris-Sud,
University Paris-Saclay, France
CHOUAIB Salem graduated with a Ph.D. in Immunology from Pierre et Marie Curie University, Paris VI. Serve as a member of the The selection committee at René Descartes University (Paris V) from 2012-present; Serve as a member of the council “Studium of Molecular Medicine” international doctorate school in Warsaw Poland from 2005-present; Serve as a member of the Scientific Steering Committee of Aix-Marseille University from 2015-present; Serve as a member of the international council in Pasteur Institute (Tunisia) from 2014-present; Serve as a member of ITMO Cancer as the coordinator of cancer immunotherapy section from 2011-present. The current research interests: immunology of cancer, Cancer Immunotherapy, Tumor microenvironment and mechanisms of carcinoma invasion, Cell-mediated cytotoxicity and tumor resistance and Translational research in Oncology.
integrative tumour immunology; cancer immunotherapy; genetic oncology

Professor of Pathology and Immunology, Medical Director, Division of Clinical Immunopathology, University of Pittsburgh Medical Center, USA
After graduation from Moscow State Medical School in 1984, postgraduation training in clinical biochemistry and clinical immunology and obtaining his PhD degree in Immunopharmacology in 1991, Dr. Shurin joined the faculty of the Department of Pathology at the University of Pittsburgh Medical Center in 1991 where he is a Professor of Pathology and Immunology and a Director of the Division of Clinical Immunopathology. His main research interests are in the field of the tumor microenvironment, cancer-mediated immunomodulation and solid tumor innervation. Professor Shurin’s pioneering studies on the immunobiology of dendritic cells in cancer opened opportunities for developing novel therapeutic approaches that are based on the protection of dendritic cell vaccines from tumor-mediated immunosuppression and polarization. At present, Shurin’s research program focuses on the immune modulating properties of chemotherapy and modulation of the tumor microenvironmental elements, including dendritic cells, regulatory T cells, myeloid-derived suppressor cells and neuroglial cells, as well as intratumoral cytokine network. Using a combination of immunological, molecular and gene therapeutic methodologies, Dr. Shurin’s team developed innovative immunotherapeutic, chemoimmunotherapeutic and nanotherapeutic approaches for cancer, which were successfully tested in the pre-clinical settings and are under investigation in clinical trials. Professor Shurin is an author of more than two hundred peer-reviewed publications and numerous reviews, an editor of several books, including “Dendritic Cell in Cancer”, “Infection and Cancer” and “The tumor immunoenvironment” books, an organizer of several popular international conferences on Immune-Mediated Diseases, Immunodiagnostics and Immunomonitoring and Cancer Immunotherapy. He is a member of several grant review and editorial boards and many clinical and research societies.

Institute of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, UK
He studied Biochamistry at the University of Edinburgh and performed doctoral studies at the University of Warwick. He did postdoctoral research in the universitites of Aarhus, Denmark and Uppsala, Sweden. He was a member of the SCientific Staff of the National Institute of Medical Research, London (now the Crick Institute). He was appointed as Lecturer in BIochemistry in 1981, Senior Lecturer in 1988, Reader in 1996 and Professor in 2008.
Oncogenic viruses; Papillomaviruses;Virotherapy; Oncolytic viruses; Cancer gene therapy; Cancer immunotherapy
Keynote Speakers
 |
Dr. Francesco Iorio Computational Biology Research Centre of Human Technopole Short Bio Francesco Iorio is a Research Group leader in the Computational Biology Research Centre of Human Technopole (the life science institute in Milan, Italy). His group works at the interface of biology, machine learning, statistics and information theory to understand and predict how genomic alterations and molecular traits from other omics contribute to pathological processes, biological circuits’ rewiring and impact therapeutic responses in human cancers and other diseases. Their research aims to advance human health by designing algorithms, computational tools and novel analytical methods for integrating and analysing pharmacogenomics and functional-genomics datasets to identify new therapeutic targets, biomarkers and drug repositioning opportunities. The Iorio Group is contributing to creating a comprehensive map of all the genetic dependencies in human cancers and developing a computational infrastructure for translating this map into guidelines for early-stage drug development and precision medicine. They design, implement and maintain bioinformatics methods and original tools for the assessment of cancer pre-clinical models, the pre-processing, analysis and visualisation of genome-editing screening data for the in-silico correction of new-technology-specific biases in such data, and the optimisation of single guide RNA libraries for pooled CRISPR-Cas9 screens and other experimental settings. They are also interested in big-data analytics, developing biomedical predictive models based on non-biomedical data, and computationally efficient constrained randomisation strategies for testing combinatorial properties in large-scale genomic datasets and networks. |
 |
Dr. Travert Hart Short Bio Within the framework of these overarching goals, a wide array of tutorial projects are possible. Computational projects could include reanalysis of existing data to improve our ability to detect gene-gene and gene-drug interactions, enhancing the BAGEL algorithm to include other molecular data such as gene expression and copy number, or helping evolve BAGEL from a gene-centric to a pathway-centric model. Experimental tutorials could involve any aspect of a CRISPR genomic screen, from lentiviral library construction to large-scale tissue culture, screening, and sequencing, and ultimately to validation and characterization of specific targets using small molecule inhibitors, automated microscopy, and other proteomic and phenotypic assays. |
 |
Dr. Andrea Morandi Short Bio |
 |
Dr. Mariateresa Fulciniti Short Bio |
 |
Prof. Dr. Vasso Apostolopoulos Short Bio |
 |
Dr. Davide Barbagallo Short Bio |
List of accepted submissions (13)
| Id | Title | Authors | Presentation Video | Poster PDF | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sciforum-067173 | Potential Candidate Gene in Underlying Molecular Mechanism Involving in Tumorigenesis of Endometriosis-associated Ovarian Cancer (EAOC) in Asian Populations | , , | N/A | N/A |
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Molecular aberrations in Endometriosis were known to be associated with an increased risk of epithelial ovarian cancer (EOCs), especially endometrioid ovarian cancer (ENOC) and ovarian clear cell carcinoma (OCCC). Causal genetic evidence currently remains elusive. The integrated study of related prognostic markers will help to identify the tumorigenesis pathways in endometriosis-associated ovarian cancer (EAOC). The objective of this study was to gain a better understanding of the tumorigenesis mechanisms that occur in the endometriosis-associated genetic variation-progressed ovarian cancer risk. We found 172 overlapping genes that changed both at the DNA and RNA levels from the literature summary of whole exome sequencing and RNA expression from GEO databases. Furthermore, we use KEGG pathways to determine a significant pathway among the gene variants. KEGG includes 50 genes that have significant enrichment pathways. In order to find genes that are specifically expressed in ovary tissue, we prioritize genes using the GTEx database. Based on their TPKM levels, we found 7 genes that were highly expressed in ovary tissue. The genes found were COL12A1, PDGFRA, C7, MYH11, MMP2, C3, and FOS. Interestingly, MYH11 was found in each sample histotype and involved in the role of the actin cytoskeleton. Several proteins influence the migratory and metastatic phenotype of tumor cells, directly or indirectly, as well as myosin protein, suggesting an explanation of tumorigenesis progression in endometriosis to ovarian cancer. Through this analysis, it has attempted to provide variants fortified for future in vivo research through this analysis. More importantly, a comprehensive study of endometriosis-associated diseases associated with ovarian cancer may open new possibilities for identifying and treating these diseases. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-068576 | 5-Nitroindazole against Lung Cancer: A multitargeted in-silico molecular docking and Molecular Dynamics simulation study | , |

|

|
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Lung Cancer has taken over all cancers in terms of diagnosis and mortality worldwide, which is why it is on the World Health Organisation’s (WHO) priority list. As per the data reported by the WHO, cancer has caused 10 million death each year, and lung cancer alone caused 1.80 million deaths in 2020. Also, the FDA has approved almost 100 drugs against lung cancer, but it is not curable as most drugs target a single protein or block a single pathway. In this study, we screened the Drug Bank library against three major proteins- Ribosomal protein S6 kinase alpha-6, Cyclic-dependent protein kinase-2, and Insulin-like growth factor-1 of lung cancer- and identified the compound 5-Nitroindazole as a multitargeted inhibitor that potentially can treat lung cancer. For the screening, we deployed multisampling algorithms such as HTVS, SP and XP, followed by the MM\GBSA calculation, and the study was extended to molecular fingerprinting analysis, ADMET calculations and Molecular Dynamics simulation analysis to understand the complex’s stability. The docking scores against the proteins 6G77, 1AQ1 and 1K3A were -6.884 Kcal/mol, -7.515 Kcal/mol and -6.754 Kcal/mol, respectively, considered in a good scoring category. Also, the compound has shown all the values satisfying the ADMET criteria, and the fingerprint analysis has shown wide similarities. The molecular dynamics of each complex have shown a cumulative deviation of less than 2 Å, which is considered best for the biomolecules, especially for the protein-ligand complexes. The best feature of the proposed drug candidate is that it targets multiple proteins of lung cancer at the same time, the chance of developing resistance is relatively less, and it drastically can reduce the burden of the pharma industry. |
|||||||||||||||||||||||||||||||||||||||||
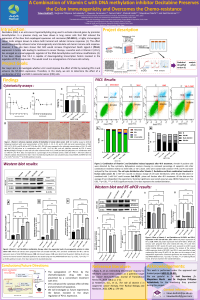
| sciforum-067497 | A combination of Vitamin C with DNA methylation inhibitor Decitabine Preserves the Colon Immunogenicity and Overcomes the Chemoresistance |
,
,
,
Fairooz Sahir ,
,
,
|
N/A |
|
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Decitabine (DAC) is an anti-cancer hypomethylating drug used to activate silenced genes by promoter demethylation. DAC induced the expression of the New York esophageal squamous cell carcinoma (NY-ESO-1), a highly immunogenic cancer testis antigen known to induce both humoral and cellular immune responses. However, DAC would increase Programmed Death Ligand-1 (PD-L1) expression in tumor cells leading to resistance to cancer therapy. Vitamin C (Vit-C) is a novel epigenetic regulator of DNA demethylation and is capable of downregulating transcription factors involved in the modulation of PD-L1 expression. Therefore, our major aim is to investigate whether Vit-C could improve the effect of DAC by reducing PD-L1 and to determine the effect of a combination of Vit-C and DAC in colorectal cancer cells. We treated the HCT-116 colon cancer cell line with DAC (20 µM) and Vit-C (1 mM) alone or in combination for 48h, then cell proliferation was assessed using CCK-8. The differential expression of immune-regulatory and pro-apoptotic markers was quantified using Western Blotting and quantitative real-time PCR. Cell apoptosis and cell cycle profile were analyzed using flow cytometry. Treatment with DAC alone induces the expression of NY-ESO-1 and upregulates the expression of PD-L1 at mRNA and protein levels compared to untreated cells. Interestingly, the combination of Vit-C with DAC decreased significantly PD-L1 expression compared to DAC alone. Further, the cytotoxic effect of DAC was primarily due to apoptosis shown by overexpression of caspase 8 and cleavage of PARP and caspase 3. This apoptotic effect was confirmed by FACS and was enhanced when Vit-C was used in combination with DAC. Vit-C prevented the upregulation of PD-L1 and enhanced the cytotoxic effect of the chemotherapeutic drug DAC. Our findings suggest Vit-C as an attractive adjuvant therapy that will promote the immune response and help to overcome immune resistance to DAC in colon cancer patients. |
|||||||||||||||||||||||||||||||||||||||||
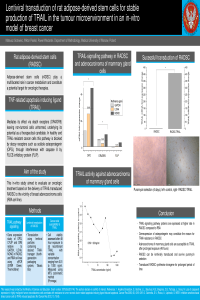
| sciforum-069082 | Lentiviral transduction of rat adipose-derived stem cells for stable production of TRAIL in the tumour microenvironment in an in-vitro model of breast cancer | , , | N/A |
|
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Adipose-derived stem cells (rADSC) play a multifaceted role in cancer metabolism and constitute a potential target for oncologic therapies. Local delivery of TNF-related apoptosis-inducing ligand (TRAIL) can limit tumour growth by selective binding to the death receptor (DR5) in cancer cells. In most healthy cells this pathway is blocked by decoy antagonists such as osteoprotegerin (OPG). This in-vitro study aimed to evaluate an oncologic treatment based on the delivery of TRAIL-transduced rADSC to the vicinity of breast adenocarcinoma cells. A plasmid containing transgene was produced using the Cloning HD system(TakaraBio) based on Plvx-EF1a-IRES-PURO plasmid and amplified TRAIL gene from the Sprague-Dawley rat kidney. Lentiviral particles were produced using VSV-G Single Shots(TakaraBio) in HEK293T cells. Cytotoxicity of TRAIL against rADSC and RBA (rat adenocarcinoma of the mammary gland) was measured using MTS Assay. Quantitative PCR and Western Blot analyses were conducted to confirm transgene overexpression and measure OPG and DR5 expression in both cell lines. Direct cytotoxicity of RBA cells to transduced rADSC was measured using exposure to a supernatant of TRAIL-expressing rADSC. Data analysis was done using the student’s t-test. Cells were successfully transduced, and TRAIL expression was 400-fold higher in transduced cells (P<0.01) than in controls. The expression of TRAIL-dependent receptors was higher in rADSC (OPG – 30-fold (P<0.01) and DR5 - 6-fold (P<0.05)). RBA cells were susceptible to exogenous TRAIL and the cell supernatant – IC50 was equal to 200 ng/ml, while rADSC were resistant to TRAIL. The results show that TRAIL-transduced rADSC are cytotoxic against adenocarcinoma cells. Overexpression of TRAIL by rADSC also stimulates their growth leading to an increased cancer microenvironment penetration. Such active surveillance based on stable local production of anticancer cytokine can limit cancer recurrence when transplanted into the vicinity of cancer tissue. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-069225 | Amelioration of Glioblastoma multiforme by the combintion of Simulated microgravity and Oncolytic viral therapy. | , | N/A |

|
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Glioblastoma multiforme (GBM) is the most common aggressive malignant primary brain tumor afflicting approximately 3.19 per 100,000 persons in the United States with an incidence 1.6 times higher in males compared to females. GBM usually arises from the glial cells known as astrocytes and it is commonly located in the supratentorial region (cortical lobes),usually affecting the frontal lobe. A unique feature of this tumor is its rapid local growth and spread making the prognosis very poor with a 5-year survival rate less than 5 %.Treatment of GBM remains challenging. Multiple therapeutic interventions are used for GBM including surgical resection of the tumor, radiotherapy and chemotherapy. Other experimental methods for the treatment of GBM include immune therapy, gene therapy, simulated microgravity therapy, and oncolytic viral therapy. We propose a combination therapy of simulated microgravity using a clinostat-based three-dimensional culture system with an oncolytic viral therapy using an autonomous rat parvovirus H-1 (H-1PV). Our hypothesis combines the beneficial effects of simulated microgravity and oncolytic viral therapy to lyse tumor cells through induction of apoptosis, decreased cell proliferation and or induction of an immune response. This proposal provides the foundations to construct novel breakthroughs in the treatment of GBM. |
|||||||||||||||||||||||||||||||||||||||||
Instructions for Authors
Submissions should be made by authors online by registering with www.sciforum.net, and using the "New Submission" function once logged into the system.
Note: Institutional email address is requested especially for the corresponding author. Please submit the abstract with the institutional email address.
Scholars interested in participating in the conference can submit their abstract (about 200–300 words) online on this website until 5th January 2023. The Conference Committee will notify the acceptance of the abstract by 31st January 2023 .
In case of acceptance, authors will be asked to submit their manuscript (short proceedings paper, 3-6 pages) before 13th February 2023. Optionally, authors of accepted abstracts will be able to submit a poster, a slides presentation (in PDF) and/or a short video presentation (max. 3-5minutes) as supporting material for the paper. Authors will receive a notification about the acceptance of their papers by 20 February 2023.
Note: All submissions will be reviewed using the powerful text comparison tool iThenticate. This procedure aims to prevent scholarly and professional plagiarism. Submissions will then be peer-reviewed by conference committees based on originality/novelty, quality of presentation, scientific soundness, interest to the readers, overall merit and English level. After the conference, all submissions will be published on sciforum.net, and only the proceeding paper (3-6 pages) will be published in the MDPI Medical Sciences Forum journal (ISSN 2673-9992).
Note: Publication of proceedings paper is free of charge.
Before publication, Medical Sciences Forum journal (ISSN 2673-9992) journal will check the plagiarism issue again. Submissions with a lack of novelty will not be published in the journal.
All participants of IECC2023 are welcome to submit an extended full paper to the conference Special Issue "IECC2023: New Targets for Cancer Therapies" of the journal Cancers, with a 20% discount on the Article Processing Charges.
Proceedings papers must be prepared in MS Word using the (see below) and should be converted to PDF format before submission. The manuscript should count at least 3 pages (incl. figures, tables and references) and should not exceed 6 pages. Carefully read the rules outlined in the 'Instructions for Authors' on the journal website and ensure that your manuscript submission adheres to these guidelines.
Manuscripts for the proceedings issue must have the following organization:
- Title
- Full author names
- Affiliations (including full postal address) and authors' e-mail addresses
- Abstract
- Keywords
- Introduction
- Methods
- Results and Discussion
- Conclusions
- (Acknowledgements)
- References
Authors are encouraged to prepare a presentation in PowerPoint or similar software, to be displayed online along with the manuscript. Slides can be prepared the same way as for any traditional conference. They should be converted to PDF format before submission.
Authors are requested to submit video presentations accompany with extended submissions. Video should be no longer than 3-5 minutes and prepared with one of the following formats: .mp4 / .webm / .ogg (max size: 250Mb). It should be submitted with the full manuscript before 16 January 2023 (full submission deadline).
Posters will be available on this conference website during and after the event. Like papers presented on the conference, participants will be able to ask questions and make comments about the posters. Posters can be presented without an accompanying proceedings paper. However, they will not be added to the proceedings of the conference.
After acceptance, please upload a copy of the proceedings/abstract as a PDF and word, in the corresponding fields, and upload the Poster PDF in the field "Presentation PDF (optional)".
1)The poster should be in PDF format
2)The minimum size for images is 148 mm × 210 mm (horizontal × vertical) at 300 dpi.
3)The content of the poster should be a comprehensive presentation of your accepted submission.
4) No copyright issues with any elements in the poster.
For detailed instructions on how to submit a poster, please contact us.
All authors must disclose all relationships or interests that could inappropriately influence or bias their work. This should be conveyed in a separate "Conflict of Interest" statement preceding the "Acknowledgments" and "References" sections at the end of the manuscript. If there is no conflict, please state "The authors declare no conflict of interest." Financial support for the study must be fully disclosed under the "Acknowledgments" section.
MDPI, the publisher of the Sciforum.net platform, is an open access publisher. We believe that authors should retain the copyright to their scholarly works. Hence, by submitting a communication paper to this conference, you retain the copyright of your paper, but you grant MDPI the non-exclusive right to publish this paper online on the Sciforum.net platform. This means you can easily submit your paper to any scientific journal at a later stage and transfer the copyright to its publisher (if required by that publisher).
Event Awards

 Winner Announcement
Winner Announcement
On behalf of the chairs of IECC 2023, we are pleased to announce the winner of the Best Paper Award:
sciforum-069265, "Novel therapeutic approaches for kras mutated lung cancer involving Lztr1 genetic alteration"
Raj Sewduth, Tonci Ivanisevic, Peihua Zhao, and Anna Sablina
The Award consists of 500 CHF and a certificate.
On behalf of the chairs of IECC 2023, we are pleased to announce the winner of the Best Presentation Award:
sciforum-069402, "Multi-omics analysis of NFE2L2 mutated TCGA-Cervical Squamous Cell Carcinoma patients"
Sri Vidya Ramisetti and Akhileshwar Namani
The Award consists of 300 CHF and a certificate.
The Awards
Number of Awards Available: 1
Best Paper Award:The Best Paper Award is given to the paper judged to make the most significant contribution to the conference. There will be one winner selected for this award, the winner will receive a certificate and 500 CHF.
Best Presentation Award:The Best Paper Award is given to the paper judged to make the most significant contribution to the conference. There will be one winner selected for this award, the winner will receive a certificate and 300 CHF.
Conference Secretariat
Ms. Rainy Han
Ms. Simon You
Ms. Alethea Liu
MDPI Branch Office, Beijing
E-Mail: iecc2023@mdpi.com
S1. Genome-wide Approaches for Target Identification.
Session Chair
Dr. Francisca Vazquez, Broad Institute of MIT and Harvard, Cambridge, USA
Session Presenters
Dr. Francesco Iorio, Computational Biology Research Centre, Human Technopole, Viale Rita Levi-Montalcini, Italy; Cancer Dependency Map Analytics, Wellcome Sanger Institute, Wellcome Genome Campus, UK.
Dr. Travert Hart, Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center, USA. Department of Cancer Biology, The University of Texas MD Anderson Cancer Center, USA.
Dr. Davide Barbagallo, Department of Biomedical and Biotechnological Sciences, University of Catania, Catania, Italy
S3. Novel Immunotherapy Targets
S4. Novel Approaches for ‘Undruggable’ Targets
Session Chair
Prof. Dr. Frank McCormick, UCSF Helen Diller Family Comprehensive Cancer Center, School of Medicine, University of California, USA, NCI RAS Initiative, Cancer Research Technology Program, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, USA profiles (https://profiles.ucsf.edu/frank.mccormick#toc-id5)
S5. Overcoming Therapeutic Resistance
Session Chair
Prof. Dr. Nicola Amodio, Department of Experimental and Clinical Medicine, Magna Graecia University of Catanzaro, Italy
Session Presenters
Dr. Andrea Morandi, Department of Experimental and Clinical Biomedical Sciences, University of Florence, Italy
Dr. Mariateresa Fulciniti, Department of Medical Oncology, Jerome Lipper Multiple Myeloma Center, Dana-Farber Cancer Institute, Boston, USA. Harvard Medical School, USA.








































