
Genes 2023 - Single-Cell Genomics Moving Forward
29–31 March 2023, Barcelona, Spain
artificial intelligence, 3D Genomics, Genomics, SingleCell Analysis
- Go to the Sessions
- Event Details
Welcome from the Chairs
Dear Colleagues,
On behalf of the Organizing Committee, it is our pleasure to invite scientists, academicians, young researchers and students from all over the world to attend in person and present their research at the International Conference “Single Cell Genomics Moving Forward” which will be held as a three-day event in Barcelona, Spain, from 29 to 31 March 2023.
The Conference is initiated and sponsored by the journal Genes (Impact Factor 4.141) with the goal of establishing a regular event to provide a venue for initiating collaborations, and discuss the latest achievements and challenges in single cell biology. Importantly, we hope to inspire young scientists to join this emerging and exciting field.
We are inviting researchers, experts, students, as well as innovative businesses in any aspect of Single Cell Multi-Omics Technologies and Analyses, Artificial Intelligence in Single Cell Genomics and Spatial Genomics and Transcriptomics to attend and present their research and products. It is only through an exchange of the widest variety of research that we can offer the best forward solutions in this challenging field. Furthermore, all presenters at the conference will be encouraged to submit a full manuscript of their presentation for consideration for publication in a Special Issue of Genes.
We look forward to meeting you and discussing your exciting single cell research in the beautiful city of Barcelona!
Professor J. Peter W. Young
Conference Secretariat
Mr. Iñigo Aristizabal
Ms. Paula Navarro
Dr. Ana Sanchis
Mr. Pablo Velázquez
Email: genesconf@mdpi.com
Event Chairs

Emeritus Professor of Molecular Ecology, Department of Biology, University of York, UK

Department of Biochemistry and Molecular Medicine, George Washington University, Washington, USA
I completed my PhD in Human Genomics in Medical University of Sofia, Bulgaria. After my postdoctoral training in Cancer Genomics (NICHD, NIH, Bethesda, MD, USA), where I led the effort for the identification of new genes responsible for human genetic disorders, in September 2013 I joined the George Washington University, as a Co-Director of the McCormick Genomic and Proteomic Center (MGPC) at the School of Medicine and Health Sciences. In this position, I focused on developing novel methods to analyze functional genetic variants through integrating layers of genomic information. In the last several months I focused my research on single cell RNA sequencing (scRNA-seq). Working on scRNA-seq data, I am particularly interested in developing novel approaches to assess genetic variation, splicing and posttranscriptional modifications, and to inter-connect them with gene expression and other phenotype features. I have over 25 year of experience with both wet and dry lab genetic approaches and technologies, including design and development of new methods and analyses.
Keynote Speakers

Head, Division Computational Genomics and Systems Genetics, German Cancer Research Center (DKFZ), Germany

Department of Statistics, Division of Biostatistics, School of Public Health
University of California, Berkeley, USA
Learning from Data in Single-Cell Transcriptomics
Sandrine Dudoit is Professor and Chair of the Department of Statistics and Professor in the Division of Biostatistics, School of Public, at the University of California, Berkeley. Professor Dudoit's methodological research interests regard high-dimensional inference and include exploratory data analysis (EDA), visualization, loss-based estimation with cross-validation (e.g., density estimation, classification, regression, model selection), and multiple hypothesis testing. Much of her methodological work is motivated by statistical inference questions arising in biological research and, in particular, the design and analysis of high-throughput microarray and sequencing gene expression experiments, e.g., single-cell transcriptome sequencing (RNA-Seq) for discovering novel cell types and for the study of stem cell differentiation. She is a founding core developer of the Bioconductor Project (http://www.bioconductor.org), an open-source and open-development software project for the analysis of biomedical and genomic data. Professor Dudoit is a co-author of the book Multiple Testing Procedures with Applications to Genomics (2008) and a co-editor of the book Bioinformatics and Computational Biology Solutions Using R and Bioconductor (2005). She was named Fellow of the American Statistical Association in 2010, Elected Member of the International Statistical Institute in 2014, and Fellow of the Institute of Mathematical Statistics in 2021. Professor Dudoit obtained a Bachelor's degree (1992) and a Master's degree (1994) in Mathematics from Carleton University, Ottawa, Canada. She first came to UC Berkeley as a graduate student and earned a PhD degree in 1999 from the Department of Statistic. She was a postdoctoral fellow at the Mathematical Sciences Research Institute (MSRI), Berkeley and in the laboratory of Professor Patrick O. Brown, Department of Biochemistry, Stanford University (1999–2001).

Josep Carreras Leukemia Research Institute, Badalona, Barcelona, Spain
Using Single Cell Multiomics in Cancer: From Knowledge to Applications
Manel Esteller (Sant Boi de Llobregat, Barcelona, Catalonia, Spain, 1968) graduated in Medicine from the Universidad de Barcelona in 1992, where he also obtained his Ph.D. degree specialising in molecular genetics of endometrial carcinoma, in 1996. He was an Invited Researcher at the School of Biological and Medical Sciences at the University of St. Andrews, (Scotland, UK) during which time his research interests focused on the molecular genetics of inherited breast cancer. From 1997 to 2001, Esteller was a Postdoctoral Fellow and a Research Associate at the Johns Hopkins University and School of Medicine, (Baltimore, USA) where he studied DNA methylation and human cancer. His work was decisive in establishing promoter hypermethylation of tumour suppressor genes as a common hallmark of all human tumours. From October 2001 to September 2008 Manel Esteller was the Leader of the CNIO Cancer Epigenetics Laboratory, where his principal area of research were the alterations in DNA methylation, histone modifications and chromatin in human cancer. Since October 2008 until May 2019, Dr Esteller was the Director of the Cancer Epigenetics and Biology Program (PEBC) in Barcelona. He is the Director of the Josep Carreras Leukaemia Research Institute (IJC), Chairman of Genetics in the School of Medicine of the University of Barcelona, and an ICREA Research Professor. His current research is devoted to the establishment of the epigenome and epitranscriptome maps of normal and transformed cells, and the development of new epigenetic drugs for cancer therapy. Website: http://www.carrerasresearch.org/en
Invited Speakers

Neurodevelopmental system biology lab, SV School-Brain Mind Institute, Swiss Federal Institute of Technology Lausanne (EPFL), Switzerland
Spatiotemporal transcriptomic mapping of the mouse embryonic brain during normal and perturbed development.
Dr. Antonio Herrera studied Pharmacy at the University of Granada, Spain, and received his Ph.D. in Biotechnology from the University of Barcelona, Spain. After that, he was awarded a “Juan de la Cierva” fellowship to start his postdoctoral training at Sebastian Pons Lab, at the IBMB Barcelona. During this time, he discovered novel mechanisms required to maintain apical-basal polarity during spinal cord development. Since August 2021 he is part of La Manno Lab at EPFL, Switzerland. His current research is focused on the generation of a detailed cellular and molecular map of the murine embryonic brain using single-cell RNA sequencing and spatially resolved transcriptomic technologies.

Director of the EPSRC Hub for Quantitative Modelling in Healthcare,
College of Engineering, Mathematics and Physical Sciences, Living Systems Institute, University of Exeter, UK
My main research interests fall within the area of biomathematics. The ultimate goal is to be able to propose novel applications of mathematics to enable the development of quantitative methods for healthcare and healthcare technologies. My current and future research plans predominantly focus on projects that could make a difference to experimental scientists and clinicians, and potentially society through the applications of mathematics for personalised prediction and decision in prevention, diagnosis or treatment of health related conditions. I am always open to hearing from prospective PhD students and early career researchers who are genuinely interested and passionate to work in an interdisciplinary setting within the area of biomathematics and applications to healthcare.

IGBMC - CNRS - University of Strasbourg, France.
Biophysics-informed interpretable deep learning to model gene regulation from single-cell and single-molecule genomic data.
Nacho Molina obtained his PhD in computational biology at the Biozentrum in Basel under the supervision of Prof. Erik van Nimwegen. After a postdoc at the EPFL in Lausanne working with Prof. Felix Naef, he obtained a Chancellor’s fellowship at the University of Edinburgh to become an independent researcher. Currently, he leads the group stochastic systems biology at the IGBMC in Strasbourg. The Molina team develops stochastic and large-scale models of eukaryotic gene regulation. The work of the team lays at the interface between machine learning and biophysics combining tools and methods from both fields to analyze and integrate single-cell genomic and imaging data.

Computational analysis of cell atlases across species
Irene Papatheodorou leads the Gene Expression team, which focuses on gene expression analyses at tissue and single cell level across species. Her team delivers tools and services for the submission, archiving, analysis and visualisation of functional genomics experiments. After completing her degree in Genetics at University College London, Irene focused on Bioinformatics through her MSc and PhD studies at Imperial College.

Department of Clinical Microbiology and Immunology, Sackler Faculty of Medicine, Tel Aviv University, Israel.
Dissecting the Immune-Controlled Signalling Networks During Tissue Development and Cancer
Dr. Cohen is the head of Systems Immunology & ImmunoGenomics Laboratory, specialized in Developmental-immunology and Cancer-Immunology. She holds a Ph.D. in ‘Neuro-Immunology’ from the Weizmann Institute of Science. She then spent four years as a Postdoctoral Fellow at the Weizmann Institute of Science working on ImmunoGenomics. In 2019, she conducted a second Postdoctoral Fellowship at the Department of Oncological Sciences, Icahn School of Medicine Mount-Sinai, NYC in ‘Tumor-Immunology’. www.mcohenlab.com

Department of Biochemistry and Molecular Medicine, George Washington University, Washington, USA
I completed my PhD in Human Genomics in Medical University of Sofia, Bulgaria. After my postdoctoral training in Cancer Genomics (NICHD, NIH, Bethesda, MD, USA), where I led the effort for the identification of new genes responsible for human genetic disorders, in September 2013 I joined the George Washington University, as a Co-Director of the McCormick Genomic and Proteomic Center (MGPC) at the School of Medicine and Health Sciences. In this position, I focused on developing novel methods to analyze functional genetic variants through integrating layers of genomic information. In the last several months I focused my research on single cell RNA sequencing (scRNA-seq). Working on scRNA-seq data, I am particularly interested in developing novel approaches to assess genetic variation, splicing and posttranscriptional modifications, and to inter-connect them with gene expression and other phenotype features. I have over 25 year of experience with both wet and dry lab genetic approaches and technologies, including design and development of new methods and analyses.

Dosage compensation dynamics unmasked by allele-specific single-cell transcriptomics
Reinius Lab: http://www.reiniuslab.com/News

Multimodal methods in single cell immunogenomics
Systems Genomics - Heinäniemi Lab: https://uefconnect.uef.fi/en/group/systems-genomics-heinaniemi-lab/

Somatic mutations in cancer and normal tissues
Abel Gonzalez-Perez obtained his PhD in Bioinformatics from the University of Havana in 2006. He is currently a Research Associate within Nuria Lopez-Bigas lab, at IRB Barcelona. For the past 11 years, he has worked on cancer genomics, specifically on applying bioinformatics to the research of molecular mechanisms underlying mutagenesis and tumorigenesis.
Event Committee

Institute of Pathology, University Medical Center Göttingen, Germany

Department of Agronomy and Horticulture
Center for Plant Science Innovation
Nebraska Center for Biotechnology
University of Nebraska-Lincoln, USA



Department of Computer Informatics, Sofia University "st. Kliment Ohridski", Sofia, Bulgaria
I completed my PhD in Computer Science in Sofia University, Bulgaria which is related to effective data representation and transfer. Since 2006 I have been teaching in a wide variety of classes in the field of programming and theoretical computer science. My major interest is data structures and algorithms, high performance computing and its application in bioinformatics. For two years now I have been the host of a seminar related to Bioinformatics. https://fmi.uni-sofia.bg/en/faculty/petr-ruslanov-armyanov
Detailed Program
Wednesday 29 March 2023 |
||
|
13:00 – 13.30 |
Registration Desk Open (Check-in) |
|
|
13:30 – 13:45 |
Welcome from the Chairs |
|
|
Session 1. Part I |
||
|
13:45 – 14:25 |
Keynote Talk: Manel Esteller - Using Single Cell Multiomics in Cancer: From Knowledge to Applications |
|
|
14:25 – 14:40 |
Silvia Ogbeide - Single-cell multi-omics profiling in the study of colorectal cancer evolution |
|
|
14:40 – 14:55 |
Francesca Nadalin - Single-cell multi-omic lineage tracing uncouples tumour initiation and drug tolerance in breast cancer |
|
|
14:55 – 15:10 |
Enrico Sebastiani - An Investigation of the origin of neuroblastoma with single-cell transcriptomic analyses |
|
|
15:10 – 15:25 |
Igor Filippov - Single-cell re-analysis of human transcriptomes reveals shared characteristics of immune ageing |
|
|
15:25 – 15:50 |
Invited Talk:Anelia Horvath - Identification and analysis of cell-specific expressed genetic variants from scRNA-seq data | |
|
15:50 – 16:20 |
Coffee Break |
|
|
Session 2. Part I |
||
|
16:20 – 16:50 |
Invited Talk: Nacho Molina - Biophysics-informed interpretable deep learning to model gene regulation from single-cell and single-molecule genomic data | |
|
16:50 – 17:05 |
Kyoung Jae Won - Legible Gene Regulatory Network Reconstruction using Multivariate Transfer Entropy Over Single Cell Transcriptomics Data |
|
|
17:05 – 17:20 |
Bogac Aybey - Immune cell type signature discovery and random forest classification for analysis of single cell gene expression datasets |
|
|
17:20 – 18:00 |
Keynote Talk: Sandrine Dudoit - Learning from Data in Single-Cell Transcriptomics |
|
|
20:00 |
Conference Dinner |
|
Thursday 30 March 2023 |
||
|
Session 1. Part II |
||
|
08:30 – 09:00 |
Invited Talk: Merja Heinäniemi - Multimodal methods in single cell immunogenomics |
|
|
09:00 – 09:15 |
Kalle Rytkönen - Gene regulatory network analysis of decidual stromal cells and natural killer cells |
|
|
09:15 – 09:30 |
Alexandre Gaspar-Maia - Single cell multiomic analysis of Clonal Hematopoiesis reveals enhancer deregulation and increased COVID-19 inflammation severity |
|
|
09:30 – 09:45 |
Sara Terzoli - Expansion of memory Vδ2 T cells following repeated exposure to mRNA SARS-CoV-2 vaccination |
|
|
09:45 – 10:00 |
Alejandro Jiménez Sánchez - Optimized tumor metastatic phenotypes among distal metastatic lesions in pancreatic ductal adenocarcinoma |
|
|
10:00 – 10:30 |
Invited Talk: Merav Cohen - The Functional Role of Immune-Related Intercellular Signaling Networks during Tissue Development and Cancer |
|
|
10:30 – 11:00 |
Coffee Break sponsored by Parse Biosciences |
|
|
Session 1. Part III |
||
|
11:00 – 11:15 |
Enrico Gaffo - Benchmarking of methods for assessing circular RNA differential expression in single-cell RNA-seq data |
|
|
11:15 – 11:30 |
Amy Hamilton - Unlocking the potential for single-cell sequencing at scale with combinatorial indexing |
|
|
11:30 – 11:45 |
Celia Alda Catalinas - Mapping the functional impact of immune disease associated regulatory elements through single-cell CRISPR-based screens |
|
|
11:45 – 12:00 |
Rapolas Zilionis - Robust and versatile ultra-high-throughput single-microbe genome sequencing |
|
|
12:00 – 12:15 |
Veredas Coleto Alcudia - TOTEM: A Web Tool For Tissue Enrichment On Gene Lists With Single Cell Resolution |
|
|
12:15 – 12:30 |
Emma Busarello - Harmonizing the annotation of single cells in normal and aberrant hematopoiesis with the Cell Marker Accordion |
|
|
12:30 – 13:45 |
Lunch |
|
|
Session 3 |
||
|
13:45 – 14:00 |
Kyoung Jae Won - Image processing approach to spatial genomics data identifies tissue architecture and cell-contact specific gene regulation |
|
|
14:00 – 14:15 |
Biao Lu - Alteration of transcriptions loaded in engineered extracellular vesicles using transmembrane scaffolds |
|
|
14:15 – 14:30 |
Susmita Datta - Inferring cell-cell communications from spatially resolved transcriptomics data |
|
|
14:30 – 14:45 |
Alba Garrido Trigo - Macrophage and neutrophil heterogeneity at single-cell spatial resolution in inflammatory bowel disease |
|
|
14:45 – 15:00 |
Kang Jin - Gene colocalization-aware segmentation of image-based spatial transcriptomics using Graph Neural Networks |
|
|
15:00 – 15:30 |
Cofee Break |
|
|
Session 1. Part IV |
||
|
15:30 – 16:00 |
Invited Talk: Björn Reinius - Dosage compensation dynamics unmasked by allele-specific single-cell transcriptomics |
|
|
16:00 – 16:15 |
Evgenia Ulianova - Cell-Level Analysis of Telomeric Transcripts and Telomere-Associated Gene Variants |
|
|
16:15 – 16:30 |
Meltem Omur - Characterization of gene regulatory networks and combinatorial transcription factor interactions during pancreatic β-cell differentiation |
|
|
16:30 – 16:45 |
Florina Moldovan - Estrogen Regulation of POC5 centriolar protein by ERafla is Altered in Human Scoliotic Cells |
|
|
16:45 – 17:00 |
Efthalia Preka - Dynamic transcriptomic alterations of microglia following cranial irradiation in the juvenile mouse hippocampus |
|
|
17:00 - 17:30 |
Invited Talk: Antonio Herrera Camacho - Spatiotemporal transcriptomic mapping of the mouse embryonic brain under folate-deficient conditions |
|
Friday 31 March 2023 |
|
|
Session 2. Part II |
|
|
08:30 – 09:10 |
Keynote Talk:Oliver Stegle - From genotype to phenotype with single-cell resolution |
|
09:10 – 09:25 |
Piotr Słowiński - Model-driven AI for multi-omics data analysis |
|
09:25 – 09:40 |
Yunhee Jeong - scMaui: variational autoencoders combined with adversarial learning reveal cellular heterogeneity from single-cell multiomics data and handle multiple batch effects independently |
|
09:40 – 10:10 |
Invited Talk: Krasimira Tsaneva Atanasova - Topological data analysis for single cell sequencing data |
|
10:10 – 10:40 |
Coffee Break |
|
Session 1. Part V |
|
|
10:40 – 11:10 |
Invited Talk: Irene Papatheodorou - Computational analysis of cell atlases across species |
|
11:10 – 11:20 |
Karin Engström - Comparison between scRNA-seq and snRNA-seq as sequencing strategies in five different tissues. |
|
11:20 – 11:35 |
Martina Höckner - Single-cell analysis reveal Cd-related effects on immune cells (coelomocytes) from the earthworm Lumbricus terrestris |
|
11:35 – 11:50 |
Ana Hernández de Sande - Cell-type-specific characterization of miRNA gene dynamics in immune cell subpopulations during aging and atherosclerosis disease progression at single-cell resolution |
|
11:50 – 12:05 |
Nadine Bestard Cuche - Marked regional heterogeneity of white matter glia in the human frontal lobe, cerebellum and spinal cord |
|
12:05 – 12:20 |
Oksana Ivanova - LMNA R482L mutation-specific impairments of skeletal muscle metabolism are associated with pathological accumulation of reactive oxygen species |
|
12:20 – 12:50 |
Invited Talk: Abel González -Somatic mutations in cancer and normal tissues |
|
12:50 |
Award Ceremony and Closing Remarks |
Conference Book

Conference Dinner

Wednesday 29 March, 20:00h
40 EUR per person
We invite you to join us at the Conference Dinner at Abrassame, a cutting-edge restaurant specialised in Mediterranean cuisine which, in addition to its location at the pictouresque terrace of Arenas de Barcelona, will make of your evening at the restaurant an experience to remember.
The dinner will be at an additional cost of 40 EUR and will need to be booked separately in an independent registration. Please register and pay for it here before 19 March 2023 and save your seat. You are welcome to bring any accompanying persons by booking their seats. Please note that registrations onsite will not be permitted.
 |
 |
 |
Registration
The registration fee includes attendance of all conference sessions and morning/afternoon coffee breaks.
Participation to the conference is considered final only once the registration fees have been paid. The number of participants is limited: once the number of paid registrations reaches the maximum number of participants, unpaid registrations will be cancelled.
Please note that, in order to finalize the scientific program in due time, at least one registration by anyone of the authors, denoted as Covering Author, is required to cover the presentation and publication of any accepted abstract. Covering Author registration deadline is 24 February 2023. Your abstract will be withdrawn if your registration is not complete by this date.
When registering, please provide us with your institutional email address. This will accelerate the registration process. If you are registering several people under the same registration, please do not use the same email address for each person, but their individual institutional email addresses. Thank you for your understanding.
Certificate of Attendance: Participants of the event will be able to downlod an electronic Certificate of Attendance by accessing their dashboards on Sciforum.net once the event is concluded. The certificates will be found under "My Certificates" category.
|
Early Bird Until 17 February 2023 |
Supported documents | |
|---|---|---|
| Academic | 350.00 EUR | |
| Student | 200.00 EUR |
Scanned copy or photograph of your current student ID (not expired) is required. |
| Guest Editors/Board Members of Genes | 250.00 EUR | |
| Author/Reviewer of the journal Genes | 300.00 EUR | |
| Non-Academic | 500.00 EUR |
| Start date - End date | Price | |
|---|---|---|
| Conference Dinner | ...-29 March 2023 | 40.00 EUR |
Free Registration Options
| Invited Speakers and MDPI Guests |
Cancellation policy
Cancellation of paid registration is possible under the terms listed below:
|
> 2 months before the conference |
Full refund but 60 EUR are retained for administration |
|
> 1 month before the conference |
Refund 50% of the applying fees |
|
> 2 weeks before the conference |
Refund 25% of the applying fees |
|
< 2 weeks before the conference |
No refund |
Visa Support Letters
- Applicants must have paid for registration and submitted an abstract in order to get a letter of support.
- Applicants must provide us with a scan of their valid in date passport that contains a photo of them.
- Applicants must provide us with an academic CV, two references from their institution (contact information including institutional email and phone) and a letter of support from their institution to confirm that they support the delegate attending the meeting.
- This must be carried in good time before the meeting, “last minute” requests will not be processed.
VISA INFORMATION
Citizens of the EU and the EFTA
Citizens of EU and EFTA countries must present valid personal identification, an ID card or a passport. No visa is required.
Regardless of their citizenship, family members of EU and EFTA citizens must present a valid passport and they must have a specific residence permit from a Schengen country (no visa required). Otherwise, a visa must be obtained.
For more information, please visit the official website.
EU Countries: Austria, Belgium, Bulgaria, Croatia, Cyprus, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Ireland, Italy, Latvia, Lithuania, Luxembourg, Malta, Netherlands, Poland, Portugal, Romania, Slovakia, Slovenia, Spain, Sweden, United Kingdom.
EFTA Countries: Iceland, Liechtenstein, Norway, Switzerland.
Schengen Countries: Austria, Belgium, Czech Republic, Denmark, Estonia, Finland, France, Germany, Greece, Hungary, Iceland, Italy, Latvia, Liechtenstein, Lithuania, Luxembourg, Malta, Netherlands, Norway, Poland, Portugal, Slovakia, Slovenia, Spain, Sweden, Switzerland.
Citizens of the USA and Canada
No visa is required for stays of up to 90 days. However, a passport valid for at least 3 months beyond your date of departure is required (6 months recommended). Spanish government regulations may require a return or on-going ticket or proof of funds.
Other Countries
A valid passport is required and in some cases a visa must be obtained prior to entering Spain. Find your nearest Spanish embassy or consulate here.
Disclaimer
We will endeavour to present the program advertised. However, in the unlikely event that MDPI shall deem it necessary to cancel the conference, all pre-paid registration fees will be reimbursed. MDPI shall not be liable for reimbursing the cost of travel or accommodation arrangements made by individual delegates. Tours run by third parties may be subject to cancellation or alteration.
Beware of Unauthorized Registration and Hotel Solicitations
Note that Sciforum is the only official registration platform to register to Genes 2023, and that we are not associated with any hotel agency. While other hotel resellers and travel agencies may contact you with offers for your trip, they are not endorsed by or affiliated with Genes 2023 or Sciforum. Beware that entering into financial agreements with non-endorsed companies can have costly consequences.
Insurance
The organizers do not accept liability for personal accident, loss, or damage to private property incurred as a result of participation in the Genes 2023. Delegates are advised to arrange appropriate insurance to cover travel, cancellation costs, medical, and theft or damage of belongings.
Payment methods
Wire transfer, Credit card
Currencies accepted by this event
Swiss francs (CHF) , Euros (EUR) and US dollars (USD)
Instructions for Authors
The "Genes 2023: Single-Cell Genomics Moving Forward" Conference will accept abstracts only.
To register for the conference, please follow this link.
To present your research at the event
Create an account on Sciforum if you do not have one, then click on ‘New Submission’ on the upper-right corner of the window, or by clicking on ‘Submit Abstract’ at the top of this webpage. Indicate what thematic area is best suited for your research.
- Submit an abstract in English (minimum 150 words and maximum 300 words). The deadline to submit your abstract is 20 January 2023. You will be notified by 3 February 2023 about the acceptance for oral presentation.
- Upon submission, you can select if you wish to also be considered for oral presentation. Following assessment by the Chairs, you will be notified by 3 February 2023 in a separate email whether your contribution has been accepted for oral presentation.
- Please note that, in order to finalize the scientific program in due time, at least one registration by any of the authors, denoted as Covering Author, is required to cover the presentation and publication of any accepted abstract. Covering Author registration deadline is 24 February 2023. Your abstract will be withdrawn if your registration is not complete by this date.
Oral Presentations
Short talks will be 15 min long including questions (12+3 Q&A). The typical presentation should be up to 10–12-minute talk.
All presentations must be uploaded in advance. Please bring your presentation to the registration desk at least 30 min before the start of your session, or at the end of the previous day for the early morning sessions. A technician will be available to assist you.
Publication Opportunities
- All accepted abstracts will be available online in Open Access form on Sciforum.net.
- Participants of this conference are encouraged to contribute with a full manuscript to our Special Issue "Single-Cell Genomics Moving Forward" in the journal Genes.
- The conference participants will be granted a 20% discount on the publishing fees for the Special Issue.
- Genes is indexed by the Science Citation Index Expanded (Web of Science), MEDLINE (PubMed) and other databases, and has an Impact Factor of 4.141 (2021) .
Venue, Travel and Accommodation
VENUE
BARCELONA
Barcelona is the capital and largest city of Catalonia and is Spain's second largest city, with a population of over one and half million people.

Located on the northeastern Mediterranean coast of Spain, this city has a rich and diverse history, with its roots dating back to Roman times. The fruitful medieval period established Barcelona's position as the economic and political centre of the Western Mediterranean. The city's Gothic Quarter bears witness to the splendour enjoyed by the city from the 13th to the 15th centuries.
The 20th century ushered in widespread urban renewal throughout Barcelona city, culminating in its landmark Eixample district, which showcases some of Barcelona's most distinctive Catalan art-nouveau, or modernista, buildings. The Catalan Antoni Gaudí, one of the most eminent architects, designed buildings such as La Pedrera, the Casa Batlló and the Sagrada Família church, which have become world-famous landmarks.
In 1992, Barcelona gained international recognition by hosting the Olympic games which brought about a massive upturn in its tourism industry. For visitors, this has translated into the very modern, yet incredibly old city you see now in the 21st century, where new elements work to both preserve and celebrate both the city’s heritage and origins.
Barcelona is plenty of outdoor markets, restaurants, shops, museums, and churches. The city is also very walkable, with an extensive and reliable Metro system for more far-flung destinations.
For a complete overview, see wikitravel.org or visit barcelonaturisme.com.

THE AXA CONVENTION CENTRE
The AXA Convention Centre is located in a vibrant modern zone of the city, next to the main road and with easy access from the airport, as well as the urban and suburban areas.
It is part of the "L'Illa Diagonal", a modern complex which include a shopping centre, two 4-stars hotels, several offices, a sports centre, a public park and a parking with 2500 places.
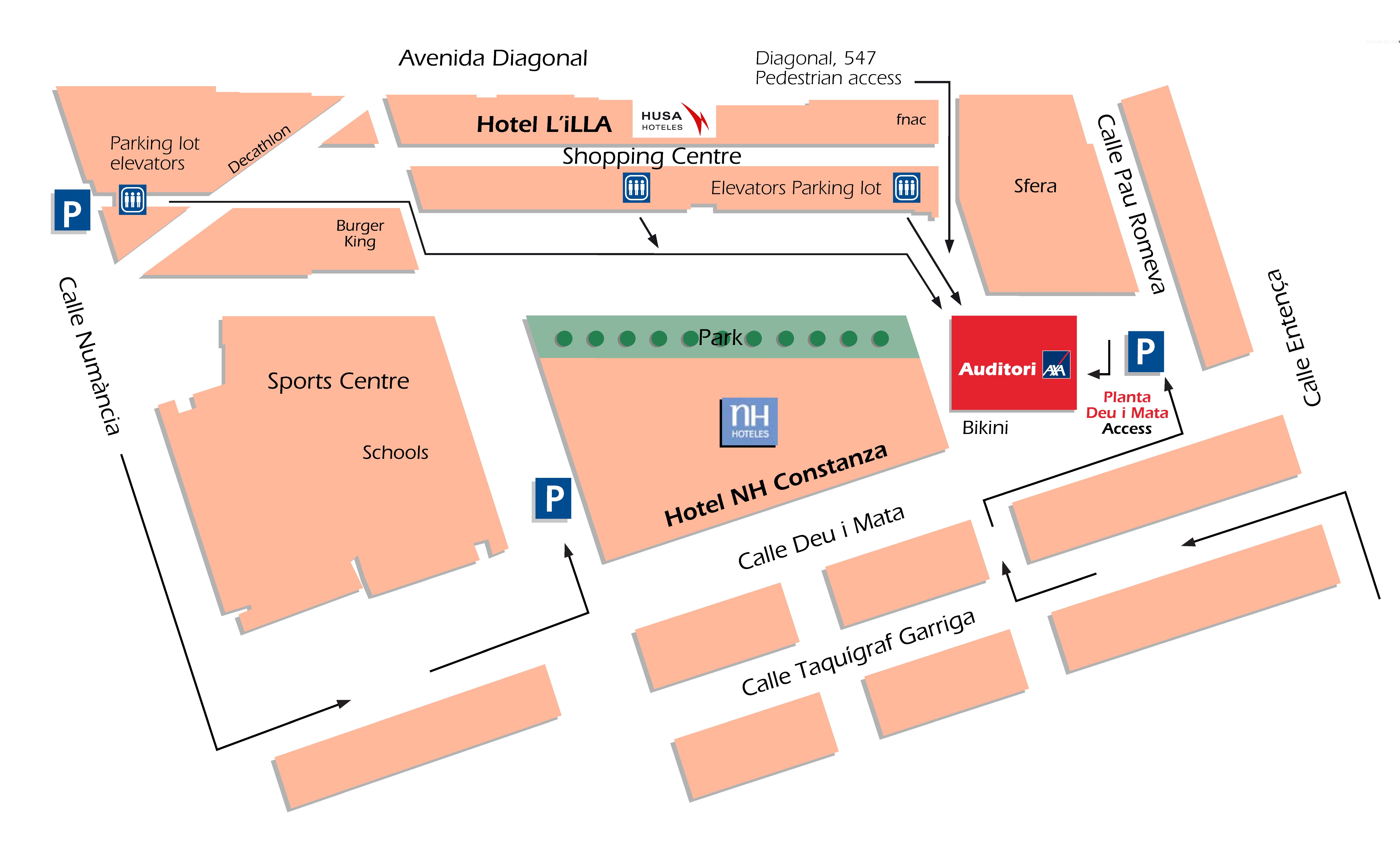
Address: Carrer Deu i Mata, 111, 08029 Barcelona (www.google.es/maps/AuditoriAxa)
TRAVEL
 BY PLANE:
BY PLANE:
The international airport of Barcelona is called Barcelona-El Prat and is located 10 km far from the city centre. The airport is well connected to airport hubs in Europe and several locations around the world. For more information about the airport and flights, please visit the official website https://www.aena.es.
Once you get to the airport, you have several options to reach the city centre and/or the conference venue.
To the city centre:
- By shuttle bus: AEROBUS (https://aerobusbarcelona.es/). This bus connects Barcelona Airport (Terminal 1 and Terminal 2) with the city center (Pl. Catalunya) in 35 minutes. It runs every day of the year with departures every 5 minutes and cost 6,75 €.
- By train: TRAIN R2 NORD (https://rodalies.gencat.cat). This train connects the Terminal 2 of the airport with the city center (Barcelona-Passeig de Gràcia). It runs every day of the year with departures every 30 minutes and cost 4,60 €.
- By taxi: taxis just outside the arrival area. The taxi from the airport to the city center (Plaza Catalunya) costs approximately 26 € during working days and 30 € during holidays, weekends and nights. Public taxis can apply four types of fares in Barcelona. These fares must be visible inside the taxi, and are usually printed on a sticker over the window. Finally, the amount payable must be indicated on the taximeter.
To the conference venue:
- By metro: LINE L9 (https://www.tmb.cat). This metro line connects Barcelona Airport (Terminal 1 and Terminal 2) with the north-west of the city (Zona Universitària). From here you can take TRAM T1, T2, T3 (get off at L'Illa stop) or bus 7, 67 (get off at Diagonal - Entença stop) (https://www.tmb.cat). The metro from the airport costs 5,15 € and the tram or bus 2,40 €. You can buy the bus/tram ticket at the tram stop or on the bus.
- By taxi: taxis just outside the arrival area. The taxi from the airport to the conference venue costs approximately 23 € during working days and 26€ during holidays, weekends and nights.
 BY TRAIN:
BY TRAIN:
Barcelona Sants station is Barcelona's largest train station and provides rail services both in and around Barcelona and for the whole of Spain and beyond. For more information about the station and train service, please visit the official website https://www.renfe.com.
Once you get to the station, you have the following options to reach the conference venue:
- By bus: LINES V7 or 78 (https://www.tmb.cat)
- By taxi: taxis just outside the station. The taxi from the station to the conference venue costs about 8 €.
ACCOMMODATION
We want your stay in Barcelona during the conference to be as good as possible. With that in mind, we have agreed a discount with several hotels to make your accommodation choice easier.
We are delighted to announce that the following hotels will offer a discounted price for all Genes 2023 attendees:
-
Arenas Atiram: The Arenas Atiram Hotel offers a highly personalized service together with very friendly, elegant facilities, and it is within walking distance from the conference venue.
-
Oriente Atiram and Meson Castilla Atiram: Located in Las Ramblas, have a fantastic central location, next to Plaza Catalunya and the famous Boqueria market, 25-30 minutes away (by metro) from the conference venue.
-
Tres Torres Atiram: Located in the center of one of the best residential neighbourhoods of Barcelona, next to the distinguished commercial and leisure districts Diagonal Avenue, 1.5 km away from the conference venue.
- Hostal Aslyp 114: The Hostal Aslyp114 is a cosy hotel with private rooms, located just 10 minutes away from the conference venue. Please add the 5% discount code ‘BCN114’ and indicate that you will be attending Genes 2023 while making your reservation.
Beware of Unauthorized Registration and Hotel Solicitations
Note that Sciforum is the only official registration platform to register to Genes2023, and that we are not associated with any hotel agency (other than the listed above). While other hotel resellers and travel agencies may contact you with offers for your trip, they are not endorsed by or affiliated with Genes2023 or Sciforum. Beware that entering into financial agreements with non-endorsed companies can have costly consequences.
VISA SUPPORT LETTERS
- Applicants must have paid for registration and submitted an abstract in order to get a letter of support.
- Applicants must provide us with a scan of their valid in date passport that contains a photo of them.
- Applicants must provide us with an academic CV, two references from their institution (contact information including institutional email and phone) and a letter of support from their institution to confirm that they support the delegate attending the meeting.
- This must be carried in good time before the meeting, “last minute” requests will not be processed.
Event Awards
In order to recognize the works presented during the conference, an award will be offered to our conference participants, which will be announced during the Award Ceremony on the last day of the conference.

The Awards
Number of Awards Available: 1
The award will be granted to the Best Presenter, selected among all non-invited talks.
S1. Single Cell Multi-Omics Technologies and Analyses
Show all accepted abstracts (27) Hide accepted abstracts (27)
List of Accepted Abstracts (27) Toggle list
S2. Artificial Intelligence in Single Cell Genomics
Show all accepted abstracts (7) Hide accepted abstracts (7)
List of Accepted Abstracts (7) Toggle list
S3. Spatial Genomics and Transcriptomics
Show all accepted abstracts (6) Hide accepted abstracts (6)
List of Accepted Abstracts (6) Toggle list










_Wang.jpg)



















