
1st International Electronic Conference on Genes: Theoretical and Applied Genomics
Part of the International Electronic Conference on Genes series
2–30 November 2020
Metagenomics, Epigenetics, DNA and RNA Sequencing
- Go to the Sessions
- Event Details
IECGE2020 has now come to a end!
On behalf of the conference team, many thanks for joining us and we hope to see you in future editions of the conference.
Upon request, the participants of the event who have registered on Sciforum will receive an electronic Certificate of Attendance.
Awards winners
We are pleased to announce that the IECGE 2020 Best Poster Awards were granted to :
By Elena Arance *, Almudena Sabio Bonilla, Mª Yarmila Gª Iglesias, Rocío Lopez Cintas, Sara Martín Esteban, Ginesa López Torres,
Cortés Valverde, Luis Javier Martínez González
NGS screening for identification of novel pexophagy-related mutant in Arabidopsis thaliana
By Katarzyna Sieńko *, Kacper Żukowski, Kenji Yamada, Shino Goto-Yamada
Congratulations to all the winners!
Welcome from the Chair
Dear Colleagues,
It is my pleasure to invite you to join the 1st International Electronic Conference on Genes (IECGE2020) , organized by the MDPI Open-Access journal Genes (Impact Factor 3.759). This free e-conference will present the latest research related to the topics of DNA and RNA sequencing, epigenetics and metagenomics.
The last ten years have consolidated Genomics as one of the faster evolving disciplines in biology. It is used by a wide range of fields, from molecular biology to ecology. Its applications go beyond genome sequencing and transcriptomic analysis, they are used to profile communities, analyze epigenetic modifications and characterize populations. This conference wants to serve as virtual space to present scientific works and change ideas under the umbrella of this fast evolving discipline.
During the month of November, IECGE2020 will allow to share your online presentations worldwide, making your work available for the audience to read and comment, and sharing ideas on the discussed topic.
All submitted abstracts will be evaluated by the conference Scientific Committee and upon acceptance, will be published in a dedicated issue of the MDPI journal Proceedings. After the conference, all participants will be encouraged to submit a full paper to one dedicated Special Issue in Genes with a 20% discount on the article processing charges (APC).
Moreover, during the conference, a number of internationally renowned speakers will share their current state-of-the-art research, through a series of pre-recorded talks and free live-streaming webinars, which will also include a Q&A session to allow audience participation.
We hope that you will join this conference to exchange ideas, merge different areas of expertise and engage in potential successful collaborations.
Kind regards,
Dr. Aureliano Bombarely
IECGE2020 Conference Chair
Conference Secretariat
M.Sc. Iñigo Aristizabal
M.Sc. Marta Mairal
M.Sc. Paula Navarro
Dr. Ana Sanchis
Email: iecge2020@mdpi.com
 Follow the conversation on Twitter with #IECGE2020
Follow the conversation on Twitter with #IECGE2020
Conference chair

Aureliano Bombarely is an Associate Professor in the Department of Biosciences at the Universita degli Studi di Milano. His research is focused in the study of the genome evolution linked to the processes of speciation, adaptation and domestication of plant species. He has been working in several popular genome sequencing projects such as tomato, watermelon, petunia, tobacco, olive and the plant model Nicotiana benthamiana. He also works in the development of bioinformatic tools related with the analysis and management of the genomic information.
Conference Committee
Prof. Dr. Michael Hofreiter: University of Potsdam, Germany
Dr. Anelia Horvath: George Washington University, USA
Prof. Dr. Yuri A. Motorin: Université de Lorraine: Nancy, France
Prof. Dr. Silvia Turroni: University of Bologna, Italy
Prof. Dr. Klaus Wimmers: Leibniz Institute for Farm Animal Biology, Germany
Keynote Speakers

Department of Mathematics and Natural Sciences, University of Potsdam, Potsdam, Germany
"Lost lineages and ghost lineages: A paleogenomic perspective on evolution"

Department of Biochemistry and Molecular Medicine, George Washington University, Washington, USA
"Correlation of SNVs to gene expression from individual scRNA-seq datasets"
I completed my PhD in Human Genomics in Medical University of Sofia, Bulgaria. After my postdoctoral training in Cancer Genomics (NICHD, NIH, Bethesda, MD, USA), where I led the effort for the identification of new genes responsible for human genetic disorders, in September 2013 I joined the George Washington University, as a Co-Director of the McCormick Genomic and Proteomic Center (MGPC) at the School of Medicine and Health Sciences. In this position, I focused on developing novel methods to analyze functional genetic variants through integrating layers of genomic information. In the last several months I focused my research on single cell RNA sequencing (scRNA-seq). Working on scRNA-seq data, I am particularly interested in developing novel approaches to assess genetic variation, splicing and posttranscriptional modifications, and to inter-connect them with gene expression and other phenotype features. I have over 25 year of experience with both wet and dry lab genetic approaches and technologies, including design and development of new methods and analyses.

"Analysis of epitranscriptomics RNA modifications by deep sequencing"
I completed my PhD in Chemistry in the Institute of Biochemistry from the Lomonossov University of Moscow. After several postdocs in the Enzymology laboratory from Gif-sur-Yvette (France) and the Institute of Biochemistry from the Lomonossov (Moscow), I have been established as a Professor of Molecular Biology at the Université de Lorraine, Nancy, France. I have over 30 years of experience in the field of RNA modifications, from studying pseudouridine synthases and methyltransferases to the development of high-throughput approaches to identify and characterize of RNA modifications

Department of Pharmacy and Biotechnology, University of Bologna, Italy
"Shotgun metagenomics of the human gut microbiome throughout our life"
Simone Rampelli is assistant professor in Chemistry and Biotechnology of Fermentation, Dept. Pharmacy and Biotechnology, University of Bologna (Bologna, Italy). His research activity, documented by 55 publications in international peer-reviewed journals and >30 participations in national and international congresses, is mainly focused on the characterization of the compositional and functional structure of the human gut microbiome, and its impact on human health.

"Sequencing the Extremes and In-betweens: Rapid Swab-to-Sequencer Microbial Profiling from the Seafloor to the Space Station"
Sarah Stahl, M.S, is a research scientist in the NASA Johnson Space Center’s Microbiology Laboratory. She has served as a project scientist for many analog and spaceflight investigations, including the Biomolecule Sequencer flight experiment that demonstrated the first DNA sequencing aboard the International Space Station (ISS). With the goal of applying sequencing to microbial monitoring, Sarah has overseen the implementation of a culture-independent, direct swab-to-sequencer method in many extreme environments, including the ISS.

Non-invasive analysis of embryonic aneuploidies in cell free DNA.
Trained in science in the University of Valencia, Spain, Dr Carmen Rubio specialized in cytogenetic studies in human reproduction, partly in the University of Barcelona. She completed her PhD in the field of Reproductive Genetics and post-doctoral research included research in male and female meiosis at the laboratory of Drs. Patricia Hunt and Terry Hassold (Washington State University, USA). She has published more than 100 papers in the main peer-reviewed specialist journals in the field, books chapters as well as numerous lectures at conferences worldwide. Currently she is the head of research for embryonic chromosomal abnormalities at Igenomix, focused on non-invasive approaches of embryo aneuploidy.

Department of Pharmacy and Biotechnology, University of Bologna, Bologna, Italy
"Shotgun metagenomics of the human gut microbiome throughout our life"
Silvia Turroni is senior assistant professor in Chemistry and Biotechnology of Fermentation, Dept. Pharmacy and Biotechnology, University of Bologna (Bologna, Italy). Her research activity, documented by 98 publications in international peer-reviewed journals and >100 participations in national and international congresses (of which > 45 as invited speaker), is mainly focused on the characterization of the compositional and functional structure of the human gut microbiome, and its impact on human health.

Genomics Unit, Leibniz Institute for Farm Animal Biology (FBN), Dummerstorf, Germany
“Epigenetics and Fetal Programming– new opportunities in animal breeding”
Klaus Wimmers is Director of the Leibniz Institute for Farm Animal Biology (FBN), Dummerstorf, Germany, and Professor of Animal Breeding and Husbandry at the University of Rostock, Germany. He obtained a degree in Veterinary Sciences from the Freie Universität Berlin in 1989, earned his doctorate at the Humboldt Universität zu Berlin in 1996 (Dr. rer. nat.) and habilitated in 2002 at the Rheinische Friedrich-Wilhelms-Universität Bonn in animal breeding and genetics. His main fields of interest are physiological, genetic and epigenetic principles of expression and differentiation of traits related to animal welfare and resource efficiency in farm animals.
Instructions for Authors
The registration for this conference is FREE and the works selected for their presentation on the conference will be published as conference proceedings with no cost.
- Authors interested in participating within the conference can submit their abstract online on this website until 21 September 2020. The abstract submitted should be about 200-word in English - the word limits are minimum 150 words and maximum 250 words.
- The Conference Committee will notify the acceptance of the abstract before 30 September 2020.
- In case of acceptance, authors will be asked to submit a conference paper (download the word template here and latex template here), and a PDF file with a graphical explanation of the research presented in the format of a poster until 21 October 2020.
- The manuscripts and posters will be available on Sciforum for discussion and rating during the time of the conference from 02 to 30 November 2020.
- After the conference, all accepted papers will be published within a dedicated issue of the MDPI journal Proceedings. In addition, all participants will be encouraged to submit a full manuscript of their presentation for consideration for publication in a Special Issue of Genes with a special 20% discount (more details to be announced soon). The submission to the journal is independent from the conference proceedings and will follow the usual process of the journal, including peer-review, APC, etc.
1. Conference Paper
Conference papers must be prepared using the Proceedings template (word template here and latex template here, following this organization:
- Full author names
- Affiliations (including full postal address) and authors' e-mail addresses (institutional addresses preferred)
- Abstract
- Keywords
- Introduction
- Methods
- Results and Discussion
- Conclusions
- (Acknowledgements)
- References
Please carefully read the rules outlined in the 'Instructions for Authors' on the journal website and ensure that your manuscript submission adheres to these guidelines.
2. Poster Presentation
Authors are encouraged to prepare a poster to be displayed online along with the manuscript. Posters can be prepared in the same way as for any traditional conference where research results are presented and should be converted to PDF format before submission so that our process can easily and automatically convert them for online display.
All authors must disclose all relationships or interests that could inappropriately influence or bias their work. This should be conveyed in a separate "Conflict of Interest" statement preceding the "Acknowledgments" and "References" sections at the end of the manuscript. If there is no conflict, please state "The authors declare no conflict of interest." Financial support for the study must be fully disclosed under "Acknowledgments" section.
MDPI, the owner of the Sciforum.net platform, is an open access publisher. We believe that authors should retain the copyright to their scholarly works. Hence, by submitting a Conference Paper to this conference, you retain the copyright of your paper, but you grant MDPI the non-exclusive right to publish this paper online on the Sciforum.net platform. This means you can easily submit your paper to any scientific journal at a later stage and transfer the copyright to its publisher (if required by that publisher).
List of accepted submissions (11)
| Id | Title | Authors | Poster PDF | ||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
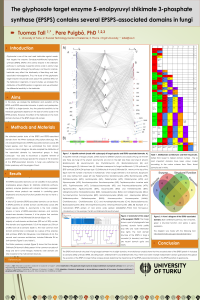
| sciforum-034483 | The glyphosate target enzyme 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) contains several EPSPS-associated domains in fungi | , |
|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
The 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS) is the central enzyme of the shikimate pathway to synthesize three aromatic amino acids in fungi, plants and prokaryotes. Glyphosate is a multi-spectrum herbicide largely utilized to control weeds, which targets the EPSPS enzyme and inhibits the production of these essential amino acids. In most plants and prokaryotes, the EPSPS protein is constituted by a single domain, whereas in fungi contains the EPSPS and several EPSPS-associated domains. Here, we perform a comprehensive analysis of 391 EPSPS proteins of fungi gathered from the Pfam database. We analyze our dataset with a bipartite graph (Cytoscape) and dollon parsimony (Count) to determine the distribution and the evolution of the 22 EPSPS-associated domains in fungi. The EPSPS-associated domains can be classified into four partially overlapping groups: shikimate pathway, other enzymes, gene expression and structural proteins. The most frequent EPSPS-associated domains are shikimate kinase, 3-dehydroquinate synthase, 3-dehydroquinate dehydratase, shikimate dehydrogenase substrate binding domain and shikimate DH. These domains are present in 56% of the proteins analyzed and 34% of proteins contain shikimate DH at the end of sequence. The most common domain architecture of the EPSPS enzyme in fungi contains 5-6 domains. A parsimony analysis suggests that a 6-domain protein is the ancestral form of the EPSPS in fungi and that alternative architectures are due to domain losses (also some gains) and duplications. The results of this study will be useful to determine the impact of glyphosate in fungi and to quantify its putative differential effects on alternative domain architectures. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-034509 |
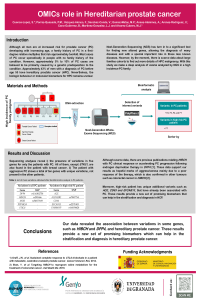
OMICs role in Heridatarian prostate cancer.
, , , , Maria del Pilar Gomez-Matas ,
Adoracion Aneas-Alaminos ,
,
,
,
Submitted: 04 Aug 2020 Abstract: Show Abstract |
,
,
,
,
Maria del Pilar Gomez-Matas ,
Adoracion Aneas-Alaminos ,
,
,
,
|
|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
Prostate cancer (PC) is one of the most prevalent tumors in the world, however, the hereditary (Hereditary PC; HPC) form is a rare pathology (ORPHA: 1331), without exceeding 6%. Despite its very low incidence, a family history of PC in a first-degree relative multiplies the risk to suffer from PC approximately twofold. Therefore, the search for genetic variables associated with detection, monitoring and treatment is unavoidable. Although the study of the genome and its expression have provided us with valuable information, it has not been able to describe biomarkers that help us resolve the appearance and evolution of the tumor. With this study, we make a deep analysis in data of exome and miRNAs analyzed by Next-generation sequencing (NGS) analysis in search of new biomarkers as variants of aggressiveness of this tumor. We performed this analysis in a family of a high incidence of PC. Our data revealed that some genes such as HIBCH and DPP4 are just present in all HPC patients. Moreover, high-risk patients have unique additional variants such as FANK1, TUBA3FP and ALDH3B2. These results provide a new set of promising biomarkers in HCP. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-034544 |
Determination of expression signature and proportion of mtDNA in plasma fractions in patients with Renal Cell Carcinoma.
, AlMUDENA SABIO BONILLA ,
Mª Yarmila Gª Iglesias ,
ROCÍO LOPEZ CINTAS ,
SARA MARTÍN ESTEBAN ,
GINESA LÓPEZ TORRES ,
CORTÉS VALVERDE ,
Submitted: 06 Aug 2020 Abstract: Show Abstract |
,
AlMUDENA SABIO BONILLA ,
Mª Yarmila Gª Iglesias ,
ROCÍO LOPEZ CINTAS ,
SARA MARTÍN ESTEBAN ,
GINESA LÓPEZ TORRES ,
CORTÉS VALVERDE ,
|

|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
Renal Cell Carcinoma (RCC) is the third most common urologic malignancy,remains one of the most lethal urological malignancies, preferably in developed countries. The incidence and mortality rates differ significantly according to sex, race, age and external factors such as smoking, obesity and hypertension increasing RCC risk. The use of novel predictive biomarkers is currently being increased as these improve the diagnosis, progression and prognosis of RCC. Since recent studies have demonstrated a promising association between mitocondrial DNA (mtDNA) copy number alteration in peripheral blood and risk of developing RCC, we conducted a case-control study to determine exosomes mtDNA content in plasma fractions as a potential novel non-invasive biomarker in liquid biopsy in order to monitor the RCC status in patients. In this way, plasma fractions highly purified in exosomes were obtained from blood samples from controls and RCC cases, and relative mtDNA content was measured by quantitative real-time polymerase chain reaction (qPCR). Our results show fragment size distribution profile and the copy numbers of nuclear regions and mitochondrial genes (in hypervariable and conserved regions) in each plasma fraction. |
|||||||||||||||||||||||||||||||||||||||||
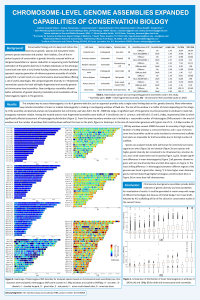
| sciforum-034647 | Chromosome-level genome assemblies expanded capabilities of conservation biology |
,
,
,
,
Erez Lieberman-Aiden ,
,
|
|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
Conservation biology aims to keep and restore biodiversity on genetic, species and ecosystem levels, prevent species extinction and protect their habitats. One of the important aspects of a conservation is genetic diversity assessed within endangered populations or species. Reduction in sequencing costs facilitated estimation of the genetic diversity in multiple individuals on the whole genome level even with a very limited budget. However, whole genome approach requires generation of reference genome assembly of suitable quality first. Current trend is to use chromosome-level assemblies offering a set of useful advantages. We compared genetic diversity in endangered species (cheetah, sea otter and others) for both old highly fragmented and recently generated chromosome-level assemblies. New contiguous assemblies allowed to calculate more precisely broadly used indicators of genetic diversity such as length and number of ROHs (runs of homozygosity) and visualize regions of low heterozygosity in the genome. In addition, we located known microsatellite loci previously used in STR-fingerprinting on chromosomes to understand diversity of what regions were estimated in previous diversity studies. Chromosome level genome assemblies provide better estimates of genetic diversity, better understanding for results of previous studies and new possibilities for visualization of results. Another new opportunity is the simple and cheap procedure for development of comprehensive STR-arrays or microarrays with known localization of each locus to use it for diversity estimates in multiple (hundreds and thousands) individuals and in low quality samples. |
|||||||||||||||||||||||||||||||||||||||||
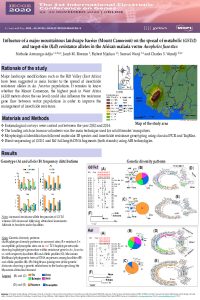
| sciforum-034654 | Influence of a major mountainous landscape barrier (Mount Cameroon) on the spread of metabolic (GSTe2) and target-site (Rdl) resistance alleles in the African malaria vector Anopheles funestus |
,
Jacob M. Riveron ,
,
,
|
|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
Increased levels of insecticide resistance in major malaria vectors such as Anopheles funestus is threatening the continued effectiveness of insecticide-based control programmes. Understanding the ecological factors impacting the spread of resistance alleles in necessary to design suitable resistance management strategies. Here we examined the influence of the highest mountain in West Africa (4,100 meters elevation) on the spread of both metabolic and target-site resistance alleles in An. funestus sensu stricto (s.s) populations. High frequencies of both 296S-Rdl (49% - 90%) and 119F-GSTe2 (67% - 81%) resistance alleles were observed An. funestus s.s. populations across Mount Cameroon. Genetic variability parameters suggested that these resistance markers underwent high levels of polymorphisms with 16 to 12 haplotypes identified respectively. However, neutrality tests were not consistent with recent expansion or resistance genes being under selection. Analysis of the maximum likelihood phylogenetic trees of haplotypes indicated that populations primarily clustered according to resistance patterns, whereas the neighbour joining trees of distances suggested that landscape variations could potentially be associated with the risk of presence and insecticide resistance for malaria vectors. These raise the need for further investigations covering different bioecological zones for a detailed report on current status of insecticide resistance in malaria vectors. |
|||||||||||||||||||||||||||||||||||||||||
Live online sessions
During the duration of the conference, a number of live online sessions will be programmed. Each session will consist on a speaker lecturing on a special topic. During each session, the participants will have the possibility to ask questions during a Q&A session. Detailed information about the topics and dates will be shared soon.
Program IECGE online sessions
PAST SESSIONS
Title Talk: Correlation of SNVs to gene expression from individual scRNA-seq datasets
Speaker: Dr. Anelia Horvath
Date and time: 3 November 2020, 3:00 PM (CET)
Title Talk: Analysis of epitranscriptomics RNA modifications by deep sequencing
Speaker: Prof. Dr. Yuri A. Motorin
Date and time: 5 November 2020, 3:00 PM (CET)
Title Talk: Non-invasive analysis of embryonic aneuploidies in cell free DNA
Speaker: Dr. Carmen Rubio
Date and time: 10 November 2020, 3:00 PM (CET)
Pre-recorded session
Title Talk: Epigenetics and Fetal Programming– new opportunities in animal breeding
Speaker: Prof. Dr. Klaus Wimmers
Date and time: 13 November 2020, 3:00 PM (CET)
Pre-recorded session
Title Talk: Lost lineages and ghost lineages: A paleogenomic perspective on evolution
Speaker: Prof. Dr. Michael Hofreiter
Date and time: 17 November 2020, 3:00 PM (CET)
Title Talk: Sequencing the Extremes and In-betweens: Rapid Swab-to-Sequencer Microbial Profiling from the Seafloor to the Space Station
Speaker: Ms. Sarah E. Stahl Rommel
Date and time: 19 November 2020, 3:00 PM (CET)
Title Talk: Shotgun metagenomics of the human gut microbiome throughout our life
Speaker: Dr. Simone Rampelli
Date and time: 25 November 2020, 3:00 PM (CET)
Live sessions content
2nd live session: Analysis of epitranscriptomics RNA modifications by deep sequencing.
Speaker: Prof. Dr. Yuri A. Motorin
Date and time: 10 November 2020
This talk is the second session of 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Prof. Yuri A. Motorin presented his most recent research related to the field of RNA modifications, from studying pseudouridine synthases and methyltransferases to the development of high-throughput approaches to identify and characterize of RNA modifications. The full recording can be found below.
1st recorded session: Non-invasive analysis of embryonic aneuploidies in cell free DNA.
Speaker: Dr. Carmen Rubio
Date and time: 5 November 2020, 3:00 PM (CET)
This talk is the third session of 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Dr. Carmen Rubio presented her most recent research as the head of research for embryonic chromosomal abnormalities at Igenomix, focused on non-invasive approaches of embryo aneuploidy. The full presentation can be found below and downloaded here.
2nd recorded session: Epigenetics and Fetal Programming– new opportunities in animal breeding.
Speaker: Prof. Dr. Klaus Wimmers
Date and time: 13 November 2020
This talk is the fourth session of 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Prof. Klaus Wimmers presented his most recent research in physiological, genetic and epigenetic principles of expression and differentiation of traits related to animal welfare and resource efficiency in farm animals. The full recording can be found below.
3rd IECGE live session: Lost lineages and ghost lineages: A paleogenomic perspective on evolution.
Speaker: Prof. Dr. Michael Hofreiter
Date and time: 17 November 2020, 3:00 PM (CET)
This talk is the thirdsession of 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Prof. Dr. Michael Hofreiter discussed the work from his group on how studying permafrost DNA could help to identify and study lost lineages of species, and establish the genetic basis of animal adaptations and the effect of environmental changes on both neutral and adaptive genetic diversity. The live session was offered via Zoom and required registration to attend. The full recording can be found below.
4th IECGE live session: Sequencing the Extremes and In-betweens. Rapid Swab-to-Sequencer Microbial Profiling from the Seafloor to the Space Station.
Speaker: Ms. Sarah E. Stahl Rommel
Date and time: 19 November 2020, 3:00 PM (CET)
This talk is the fourth live session of the 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Ms. Sarah E. Stahl Rommel, a microbiologist from the NASA Johnson Space Center discussed her work on sequencing techniques for microbial monitoring, and the implementation of a culture-independent, direct swab-to-sequencer method in many extreme environments, including the International Space Station. The live session was offered via Zoom and required registration to attend. The full recording can be found below.
5th IECGE live session: Shotgun metagenomics of the human gut microbiome throughout our life.
Speaker: Dr. Simone Rampelli
Date and time: 25 November 2020, 3:00 PM (CET)
This talk is the last live session of the 1st International Electronic Conference on Genes: Theoretical and Applied Genomics. Dr. Simone Rampelli, a researcher from University of Bologna discussed his work on the characterization of the compositional and functional structure of the human gut microbiome, and its impact on human health. The full recording can be found below.
To stay updated on upcoming live sessions on Genes organized by MDPI, be sure to sign up for our newsletter by clicking on “Subscribe” at the top of the page.
Event Awards
We are pleased to announce the availability of awards for researchers who plan to attend the IECGE2020 conference.
Applications and nominations will be assessed by Scientific Committee. Winners will be announced online after the conference.


The Awards
Number of Awards Available: 1
Award winner will receive 300 CHF and will be able to publish the extended version of one paper in the Journal Genes, free of charge (being the paper the one submitted to the conference, or another one). The extended version will have a normal peer-review procedure.Number of Awards Available: 1
Winners will be selected by the Scientific Committee after evaluation of all the posters presented during the conference. The award will consist of 200 CHF.S2. Applications of Genomic Technologies
Keynote speakers:
Prof. Dr. Michael Hofreiter
Prof. Dr. Klaus Wimmers
Prof. Dr. Silvia Turroni
Show all published submissions (11) Hide published submissions (11)
Submissions
List of Papers (11) Toggle list




















