
The 2nd International Electronic Conference on Biomolecules
Biomacromolecules and the Modern World Challenges
Part of the International Electronic Conference on Biomolecules series
1–15 November 2022
proteins, carbohydrates, nucleic acids, Lipids, Ecology, Health, Technology
- Go to the Sessions
- Event Details
IECBM 2022 is closed. Thanks for your participation.
You could download your certificate from your sciforum account.
See you in the next edition.
Welcome from the Chair
Dear Colleagues,
It is with great enthusiasm that we announce the 2nd International Electronic Conference on Biomolecules: Biomacromolecules and the Modern World Challenges (IECBM2022). The conference is organized by the MDPI open-access journal Biomolecules (Impact Factor 6.064), and will be held online from 1-15 November 2022.
This conference will provide leading scientists working in the field with an online platform on which to share their latest research and engage in exciting discussions. The main topics and sessions of the conference are:
Biomacromolecules: Proteins;
Biomacromolecules: Carbohydrates;
Biomacromolecules: Nucleic Acids;
Biomolecules: Lipids.
This online event will bring together researchers from all over the world with no concerns of travel or other related expenditures. Participation and “attendance” to the IECBM2022 are FREE of charge.
The conference invites submissions for abstracts that will be reviewed by the conference committee. The authors of accepted contributions will be invited to submit a conference paper along with a slide or poster presentation of their work. All participants will have the opportunity to examine, explore, and critically engage with research findings published in Sciforum during the conference.
After the conference, all accepted papers will be published in the proceedings of this e-conference within a dedicated issue of the MDPI journal Biology and Life Sciences Forum. In addition, all participants will be encouraged to submit a full paper to one dedicated Special Issue in Biomolecules with a 20% discount on the article processing charges (APC).
We hope that you will join this e-conference to exchange ideas, start fruitful collaborations, and make this edition a success.
Kind regards,
Prof. Dr. Vladimir N. Uversky, PhD, DSc, FRSB, FRSC
List of accepted submissions (53)
| Id | Title | Authors | Presentation Video | Poster PDF | |||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sciforum-059129 | Ohmic heating effect on the bioaccessibility of proteins from ohmic-heated nixtamalized tortillas |
Vega-Vazquez Lucia ,
,
,
|
N/A | N/A |
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
This research aimed to assess protein bioaccessibility of traditionally (TN) or ohmic heating (OH)-nixtamalized sorghum tortillas using two sorghum varieties (82w21/8133) processed at several conditions (110/120 V, 85/90 ºC). The 82w21 variety (120 V/85 ºC) displayed the highest yield (1.82 kg tortilla/kg masa) and the best sensory parameters (rollability/puffiness). A higher tannin decrease (-27.77%) was obtained compared to TN. The highest protein bioaccessibility (58.23 %) was found for OH-tortillas at 60 min in the digestible fraction, while TN showed the highest permeation rates. Concluding, OH is an environmentally friendly procedure to obtain nixtamalized sorghum flours to manufacture highly-bioaccessible protein tortillas |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-059289 | Effects of extrusion cycles on the formation of type 3 resistant starch |
Romero-García Monica ,
,
,
|
N/A | N/A |
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
The present work was to evaluate the formation of type III resistant starch in Hylon VII starch by the cycle’s extrusion process. The starch was subjected to three extrusion cycles. Starch viscosity, structural and thermal properties were determined. Results have shown that thermal properties had a decrease in the enthalpy, a decrease in the viscosity, and a loss in the crystallinity pattern because of the cycle’s extrusion. The type III resistant starch decreased for each extrusion cycle, due to the gelatinization process that occurred during the extrusion cycles. |
|||||||||||||||||||||||||||||||||||||||||
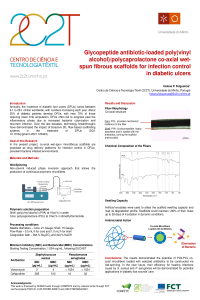
| sciforum-060119 | Glycopeptide antibiotic-loaded poly(vinyl alcohol)/polycaprolactone co-axial wet-spun fibrous scaffolds for infection control in diabetic ulcers | N/A |
|
Show Abstract |
|||||||||||||||||||||||||||||||||||||
|
Annually, the incidence of diabetic foot ulcers (DFUs) varies between 9.1 to 26.1 million worldwide, with numbers increasing each year. About 25% of diabetic patients develop DFUs, with near 70% of those requiring lower limb amputation. DFUs often fail to progress past the inflammatory phase due to increased bacterial colonization and recurrent infection. Over the last decades, technology breakthroughs have demonstrated the impact of bioactive 3D, fiber-based scaffolding systems in the treatment of DFUs (DOI: 10.1016/j.ijpharm.2021.120423). In the present project, co-axial wet-spun microfibrous scaffolds are proposed as drug delivery platforms for infection control in DFUs-prevalent bacteria infested environments. Microfibers with a shell made of poly(vinyl alcohol) (PVA) at 10wt.% in water and a core of polycaprolactone (PCL) at 10wt.% in dimethylformamide were engineered via wet-spinning in a 8wt.% Na2SO4 and 4wt.% NaOH coagulation bath. Ceftazidime and vancomycin minimum bactericidal concentrations (MBC) against S. aureus and P. aeruginosa were determined at 32.0µg/mL and 7.8µg/mL, respectively. Antibiotics were loaded at 2×MBC at the shell of the microfibers. PCL was used to sustain the scaffolding structure and convey mechanical resilience, while PVA was used primarily as a drug carrier, maintaining a local moist environment and absorbing exudates. The microfibers co-axial structure was confirmed via brightfield microscopy. Both polymers and antibiotics presence were verified via Fourier-transform infrared spectroscopy (FTIR) and UV-visible spectroscopy, following antibiotic release up to 24h in phosphate buffer (PBS, pH 7.4). Artificial exudates were used to attest the scaffold swelling capacity and map its degradation profile. Scaffolds could maintain >80% of their mass up to 28 days of incubation in dynamic conditions. Antimicrobial testing revealed the drug diffusion abilities of the scaffolding system, forming zones of inhibition in single and co-culture settings. Time-kill kinetics studies established this drug delivery platform as effective for infection control in non-sterile DFUs-mimicking environments within a 24h period. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-060269 | Coaxial wet-spun fibers loaded with AAPV – a viable option for chronic wound healing | , , , , , |

|
N/A |
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
Chronic wounds (CW) are a worldwide concern, causing serious strives on the health and quality of patients’ life. In CW, human neutrophil elastase (HNE) enzyme gets highly expressed during inflammation, reaching abnormally elevated concentrations. Additionally, prevalence of Staphylococcus aureus-induced infections remains very high and difficult to treat. Considering these phenomena, a drug delivery system made of co-axial wet-spun fibers, loaded with the tetrapeptide Ala-Ala-Pro-Val (AAPV, a known inhibitor of HNE activity) and N-carboxymethyl chitosan (NCMC, responsive to neutral-basic pH’s, characteristic of CW and endowed with antibacterial features), was proposed. AAPV was synthesized by solid-phase peptide synthesis, whereas NCMC was synthesized from low molecular weight chitosan in a chloroacetic acid mixture. HNE inhibition tests were conducted to establish the AAPV IC50 in 1.50 µg/mL and the NCMC minimum bactericidal concentration (MBC) against S. aureus in 6.40 mg/mL. These determinations were used to establish fiber loading amounts. Core-shell structures were produced with 10% w/v polycaprolactone (PCL) at the core and 2% w/v sodium alginate (SA) solutions at the shell. NCMC was mixed with SA at 2xMBC so neutral-basic pH-triggered solubility (characteristic of CW) would allow pores to be opened in the outer layer for accessing the core, where AAPV was combined with PCL. Fourier-transform infrared spectroscopy and brightfield microscopy were used to confirm the presence of the four components on the fibers and the co-axial architecture, respectively. Fibers presented maximum elongations of over 100%. Release kinetics studies conducted via UV-visible absorption spectroscopy mapped NCMC liberation overtime but were uncapable of detecting AAPV, since polymer degradation masked AAPV absorption peaks. Time-kill kinetics studies against S. aureus demonstrated the effectiveness of NCMC in eliminating this bacterium, particularly after 6 h of incubation. On its turn, AAPV guaranteed HNE inhibition. Data demonstrated the potential of SA-NCMC-PCL-AAPV co-axial systems to work as stepwise, pH-triggered delivery platforms. |
|||||||||||||||||||||||||||||||||||||||||
| sciforum-060335 | Development and characterization of calcium carbonate-quince bio-composite for pH triggered release of darfenacin hydrobromide in lower GIT: A green chemistry approach |
,
,
,
,
,
,
Muhammad Waqas
|
N/A | N/A |
Show Abstract |
||||||||||||||||||||||||||||||||||||
|
A green chemistry approach was employed to develop gastric pH resistant bio-composites for colon targeted oral delivery of darfenacin hydrobromide. The FTIR, XRD, DSC and TGA results showed good drug-polymer compatibility. The SEM images showed calcite formation in the quince hydrogel system. The drug release of 80% and 34% were observed in a phosphate buffer 6.8 pH and an acidic media, respectively. The restricted drug permeation (approx. 21.8% only) was observed through gastric membrane in an acidic media. The developed formulation significantly inhibited the development of testosterone induced prostatic hyperplasia. No organ toxicity was observed against all the developed formulations. |
|||||||||||||||||||||||||||||||||||||||||
Conference Secretariat
Ms. Sienna Tian
Ms. Billie Jiao
Ms. Yufei Shi
Ms. Sara Wang
Ms. Letty Zhu
MDPI Branch Office, Beijing
E-mail: iecbm2022@mdpi.com
Event Chair

Department of Molecular Medicine, University of South Florida, USA
Vladimir Uversky is Professor at the College of Molecular Medicine, University of South Florida. He is a biophysicist and obtained his PhD and Doctor of Science (DSc) in Physics and Mathematics at the Moscow Institute of Physics and Technology (1991) and at the Institute Experimental and Theoretical Biophysics, Russian Academy of Sciences (1998), respectively. He spent his early career investigating protein folding at the Institute of Protein Research and the Institute for Biological Instrumentation (Russia). In 1998, he moved to the University of California Santa Cruz to work on protein folding, misfolding, and protein intrinsic disorder. In 2004, he joined the Indiana School of Medicine and, in 2010, the University of South Florida, where he has since been continuing his studies on protein folding, misfolding, and intrinsically disordered proteins. Prof. Vladimir Uversky has authored over 980 scientific publications, edited numerous books, and is also serves as the editor of several scientific journals. He was included in Thomson Reuters list of Highly Cited Researchers in 2014-2020.
Event Committee

Research Director, ISOF, Consiglio Nazionale delle Ricerche, Italy
Interests: free radical chemistry; biomimetic chemistry; organic synthesis; reaction mechanism; analytical protocols for biomarkers of radical stress; oxidative DNA damage; lipid modification; fatty acid-based lipidomics

Laboratoire de Biologie des Ligneux et des Grandes Cultures (LBLGC), INRA USC1328, Université ď Orléans, France
Interests: chemistry of natural products; analytical methods; HPLC; LC-MS; polyphenols (lignans, flavonoids, phenolic acids); ethnopharmacology; history of pharmacy

Institute of Translational Medicine, Faculty of Health Sciences, University of Macau, China
Interests: novel therapeutic antibodies development; venom-based peptide & natural biomolecule prototype drugs development; cancer biomarkers & immunotherapy markers discovery for prognostic and therapeutic validation

School of Systems Biology and The Krasnow Institute for Advanced Study, George Mason University, USA
Interests: multiscale systems biology; computational biology; bioinformatics; cardiac physiology; immunology; mitochondria; cellular signaling; neuroscience; algorithms; HPC

Center for RNA Medicine, Department of Clinical Medicine, Aalborg University, Denmark
Interests: bioinformatics; cardiovascular system; circRNA; database; epitranscriptomics; lncRNA; miRNA; non-coding RNA

Department of Nutrition and Food Science, University of Granada, Campus of Cartuja, Spain
Interests: antioxidants; natural product chemistry; antioxidant activity; phytochemicals; lipids; lipid oxidation
Department of Biomedical and Molecular Sciences, Queen’s University, Kingston, ON K7L 3N6, Canada
Interests: immunology; COVID-19 and ACE2 receptor; cancer; inflammation; TLR receptor; Trk receptor; GPCR signaling; insulin receptor; glycosylation; NEU1 sialidase
CSIC Instituto de Biomedicina y Biotecnología de Cantabria (IBBTEC), Santander, Spain
Interests: RAS; ERK; MAP kinases; scaffold proteins; signalling spatial regulation
Department of Chemistry, University of Coimbra, 3004 - 535 Coimbra, Portugal
Interests: transport properties; thermodynamic properties; electrolytes; polyelectrolytes; polymers; carbohydrates; drugs; carriers; dental alloys; corrosion
Call for Abstracts
The 2nd International Electronic Conference on Biomolecules will be held from 1-15 November, 2022. IECBM aims to promote and advance the exciting and rapidly changing field of biomacromolecules and the modern world challenges. All abstracts and presentations will be held online at https://iecbm2022.sciforum.net/.
Topics of interest include, but are not limited to:
-
Biomacromolecules: Proteins (Session A);
-
Biomacromolecules: Carbohydrates (Session B);
-
Biomacromolecules: Nucleic Acids (Session C);
-
Biomolecules: Lipids (Session D).
The conference will be completely free of charge—both to attend and for scholars to upload and present their latest work on the conference platform. There will also be the possibility to submit selected papers to the journal Biomolecules ( ISSN: 2218-273X; Impact Factor: 6.064) with a 20% discount on the APCs.
IECBM 2022 offers you the opportunity to participate in this international, scholarly conference without having the concern or expenditure of travel—all you need is your computer and access to the Internet. We would like to invite you to “attend” this conference and present your latest work.
Abstracts (in English) should be submitted by 29 July 2022 online at https://iecbm2022.sciforum.net/. For accepted abstracts, the presentations can be submitted by 16 September 2022. The abstracts and presentations will be available on Sciforum for discussion during the time of the conference (1-15 November 2022) and will be then published in the Journal Biology and Life Sciences Forum.
We hope you will be able to join this exciting event and support us in making it a success. IECBM 2022 is organized and sponsored by MDPI, a scholarly open-access publisher based in Basel, Switzerland.
Instructions for Authors
Submissions should be made by authors online by registering with https://iecbm2022.sciforum.net/, and using the "New Submission" function once logged into the system.
- Scholars interested in participating in the conference can submit their abstract (about 200–300 words) online on this website until 29 July 2022.
- The Conference Committee will notify the acceptance of the abstract by 10 August 2022.
- If the abstract is accepted for this conference, the author will be invited to prepare a full description of their work in the form of a Poster/PowerPoint/Video Presentation (max. 5 minutes), until the submission deadline 9 September 2022. Authors will receive a notification about the acceptance of their presentations by 11 October 2022.
- The abstracts and presentations will be available on https://iecbm2022.sciforum.net/ for discussion and rating during the time of the conference, from 1-15 November 2022.
- All accepted abstracts+presentations will be published as one dedicated volume in MDPI Biology and Life Sciences Forum (ISSN: 2504-3900). Publication of the proceedings will be free of charge.
- The open-access journal Biomolecules (Impact Factor 6.064) will publish a conference Special Issue. Conference participants are encouraged to submit a full paper to the dedicated Special Issue and will receive a 20% discount on the Article Processing Charges (APC), in case their paper is accepted for publication after peer-review.
Submission of Manuscripts
Authors are encouraged to prepare a presentation in PowerPoint or similar software, to be displayed online along with the manuscript. Slides can be prepared the same way as for any traditional conference. They should be converted to PDF format before submission.
Authors are also encouraged to submit video presentations. This is an unique way of presenting your paper and discussing it with peers from all over the world. Video should be no longer than 5 minutes and prepared with one of the following formats: .mp4 / .webm / .ogg (max size: 250Mb). Videos should be submitted with a copy of the accepted abstract by 9 September 2022.
Posters will be available on this conference website during and after the event, participants will be able to ask questions and make comments about the posters. Authors that wish to present a poster must follow the following instructions: 1) The poster should be in PDF format; 2) The content of the poster should be a comprehensive presentation of your accepted submission; 3) No copyright issues with any elements in the poster. Posters should be submitted along with a copy of the accepted abstract by 9 September 2022.
All authors must disclose all relationships or interests that could inappropriately influence or bias their work. This should be conveyed in a separate "Conflict of Interest" statement preceding the "Acknowledgments" and "References" sections at the end of the manuscript. If there is no conflict, please state "The authors declare no conflict of interest." Financial support for the study must be fully disclosed under "Acknowledgments" section.
MDPI, the publisher of the Sciforum.net platform, is an open access publisher. We believe that authors should retain the copyright to their scholarly works. Hence, by submitting a communication paper to this conference, you retain the copyright of your paper, but you grant MDPI the non-exclusive right to publish this paper online on the Sciforum.net platform. This means you can easily submit your paper to any scientific journal at a later stage and transfer the copyright to its publisher (if required by that publisher).
Event Awards
The Sponsor Biomolecules offers one Best Contribution Award and one Best Poster Award to our participants.


The Awards
Number of Awards Available: 1
300 CHF + publication waiver in Biomolecules JournalNumber of Awards Available: 1
300 CHFTerms and Conditions:
All formats of presentations will be eligible for the Best Contribution Award.
Evaluation Criteria:
- Originality / Novelty
- Significance of content
- Scientific soundness
- Interest to the readers
- English language and style
Evaluation Process:
- Each Evaluation Committee member will assess the criteria outlined above for each application.
- Presentations will be ranked from highest to the lowest total score.
- If two or more authors get the same score, further evaluation will be carried out.
- Winners will be announced online after the conference.








































_Kwok.png)













